Abstract
This study employs computational analysis to identify and characterize liver inflammation-related genes in humans. Focusing on five genes (MAPK1, ITGA2, CDK2, IL6, and OAS2), the research includes gene sequence retrieval (NCBI), chromosomal mapping, gene structure prediction, motif analysis, protein-protein interaction networks, and gene ontology annotation. The analysis reveals that these genes are randomly distributed across chromosomes and are involved in key biological processes related to liver inflammation. Protein-protein interaction analysis identifies ITGA2 as a central gene with high interaction degrees. Gene ontology results show IL6 and OAS2 significant roles in the defense response to viruses. The findings offer insights into liver inflammation mechanisms and suggest potential biomarkers for early disease monitoring and therapeutic strategies. Further experimental validation is needed to confirm these computational predictions.
Keywords
Human, Computational Analysis and Liver Inflammation.
Introduction
Inflammatory is a typical reaction to infection and damage, whether it be local or systemic. A good immune system response adopts a certain pattern, with the first reaction being powerful but brief, ending in the prohibiting of the injury and a return to equilibrium. This distinctive inflammatory route is necessary for tissue repair and remodeling, as well as regaining a strong homeostasis and serious function (Hotamisligil and Erbay, 2008). The evolutionary advantages of an optimally functioning immune response are clear in defending against harmful invaders. Because immunological responses are connected to energy metabolism, it may be claimed that integrating these systems and collaborating to respond to variations in the energy and dietetic environment would be advantageous. To maintain a healthy homeostasis, these reactions must be managed both temporally and spatially. Chronic disturbance of metabolic balance, such as overnutrition, may result in altered immunological responses. persistent inflammatory induction, particularly in through metabolism important organs such as the liver and adipose tissue, stimulates the release of cytokines that are proinflammatory, severe phase including proteins, inflammatory lipids, and additional physiological mediators of inflammation into bloodstream, resulting in a systemic inflammation situation (Hotamisligil, 2017; Gregor and Hotamisligil, 2011; Minihane et al., 2015). These progressions play an important part in the development of long-lasting metabolic diseases such like overweightness, diabetes, fatty liver disease, and heart related disease (Hotamisligil, 2006), as well as providing the mechanism for risk factors for virus infectious disease like coronavirus disease -2019 (Stefan et al., 2021). To support treatment efforts for metabolically illnesses, it is critical to get a greater knowledge of the inflammation to anticipate the individual's homeostatic inflammatory condition. To this goal, tissue-derived plasma biomarkers that represent tissue specific and systemic inflammatory changes across time would be extremely useful. Various systemically inflammatory markers, such as C-reactivity protein (CRP), IL (interleukin) 6, Interleukin 18, fibrinogen, as well as adhesion proteins vasculature cell adherence protein 1 (VCAM-1). (Shibata et al., 2009; Pradhan, 2001; Li et al., 2009; Thorand et al., 2005; Kelesidis et al., 2006) have been shown to identify "end-stage" chronic poor-quality inflammatory diseases that include type 2 diabetes, cardiovascular illness, and carcinoma. Conversely, elevated plasma concentrations of a protein called a substance that reduces inflammation, were shown to be adversely linked with Cardiovascular (Shibata et al., 2009), type 2 diabetes (Li et al., 2009), and obesity-related malignancies (Kelesidis et al., 2006). To determine how much such and additional markers indicate the initial phases of illness development at the level of tissue rather than at the level of the system, an extensive evaluation of pro-inflammatory markers at the systemic threshold is required to select markers that allow evaluation of tissue-derived mild inflammation of low grade. The latest advances in a high-through technologies have enabled the generation, analysis, and integration of huge multiomics databases at both the cellular and molecular levels in order to uncover genetic markers of disease progression. The growing use of in computational methods and bioinformatics has motivated investigators to combine multiple datasets with prior information and databases to provide a systems-level picture of illness progression (Mcdermott et al., 2013; Vafaee et al., 2018). The analysis of gene set enrichment has emerged as the premier method for analysing omics information, decreasing intricacy and offering a systems perspective on the biochemical mechanisms associated with the progression of diseases (Khatri et al., 2012). There have been several ways offered in the research for this purpose. Most strategies rely on levels of expression and biological processes (Khatri et al., 2012; García Campos et al., 2015). The technique reported here reverses this paradigm, starting with known pathways and tissue-derived biomolecules, resulting in readily interpretable potential biomarkers that may aid in disease monitoring in the early stages and treatment regimens. By focusing on the main processes that contribute to persistent low-grade inflammation, we were able to identify common traits across a wide spectrum of metabolic illnesses. Jolanda et al. (2021) developed an in-silico technique that predicts blood-based candidate biomarkers using prior knowledge of dysregulated process in long-lasting inflammation. This technique was validated using biomarker databases, experimental data, and scientific literature to find blood-based biomarkers indicating the dynamic inflammatory state throughout the subclinical process of chronic low-grade inflammation in the liver and adipose tissue. Several researches have recently looked at the underlying processes that cause hepatic inflammation to develop. First, immune cell recruitment to the liver was thought to be critical for the onset of HBV-related hepatic inflammation and subsequently longterm persistent infection. (Zhu et al., 2017; Hou et al., 2018). The various cytokine expression levels were also discussed during different stages of HBV infection. (Trehanpati and Vyas, 2017; Li et al., 2016). For example, Interleukin-22 is a cytokine implicated in the aetiology of liver disease but has a contentious function in hepatic inflammation in HBV patients. Cobleigh and Robek (2013); Gao et al. (2013). Second, endoplasmic reticulum and mitochondrial disorders have been linked to the pathophysiology of hepatic inflammation, which leads to liver damage (Kim et al., 2017; Mansouri et al., 2018). Third, structural alterations in the gut microbes, translocation of bacteria, and subsequent immunological damage may influence the onset and progression of chronic HBV-induced liver inflammation (Yang et al., 2018).
MATERIALS AND METHODS
Sequence Retrieval
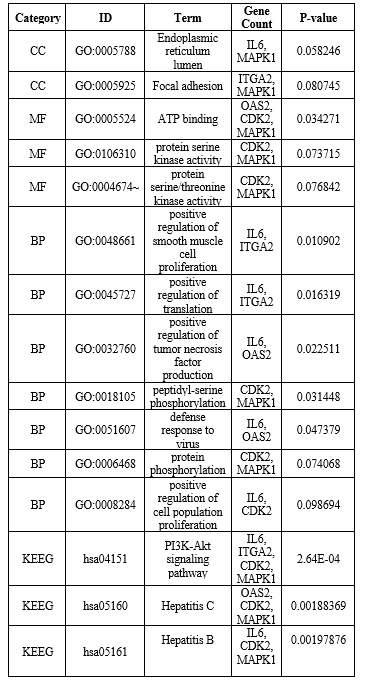
Gene bank was searched to identify liver inflammation related genes in Homo sapiens (Human). A query of 5 genes found in various articles was used for this purpose. This was also accomplished with the help of National Center for Biotechnology Information (NCBI). NCBI was used to obtain genomic sequences, protein length, chromosomal location, start and end position of each gene. Using an online tool (https://web.expasy.org/compute pi/), the isoelectric point and molecular weight of identified proteins were calculated. For further research and characterization, protein sequence data of liver inflammation related genes were created (Table 1).
Table 1: Genes characteristics.
Chromosomal mapping
For chromosomal mapping, map chart file generated included the gene symbol, chromosome number, gene position and chromosome length. Map chart file was used as input source and visualized chromosomal mapping by using bioinformatics tool (Map chart) (http://www.biometris.wur.nl/UK/Software/MapChart/download) (Voorrips, 2002). Chromosomal mapping visualized the position of each liver inflammation related genes on chromosomes.
Gene structure and Motif analysis
The coding and genome succession of all predicted genes was downloaded using NCBI. WebScipio was used to get a high-quality gene structure prediction from protein query (https://www.webscipio.org/) (Odronitz et al. 2008). In motif analysis, combined information of 5 gene sequences has been subjected to Multiple Expectation maximizations for Motif Elicitation (MEME) suit server 4.11.2 to identify the motif location and motif width. The parameters adopted for MEME analysis include number of repetitions, any; protein sequence length 309.5; maximum number of motifs, 5; and optimal breadth of every motif, among 6 and 60 residuals (Cao et al. 2021).
Protein-protein association
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) Ver. 11.0 (https://string-db.org/cgi/input.pl) was used to analyze protein-protein association networks (Kim et al. 2021). STRING programme was used to input the amino acid sequence, and the protein-protein analysis result was downloaded to reflect the gene-gene interaction and visualized via cytoscape.
Gene ontology (GO Analysis)
The bioinformatics programmed Database for Annotation, Visualization, and Integrated Discovery (DAVID) was used to do the GO annotation study (https://david.ncifcrf.gov/tools.jsp) (Huang et al., 2009; Shao et al., 2009; Jiao et al., 2012). liver inflammation related genes symbol set used as an input source for GO annotation. Consequently, functional annotation tool developed the biological process (gene/gene-product contribute in biological process), molecular function (gene product that is biochemically active), cellular components (cell area in which gene product is active) and functional category for ontologies.
RESULTS
Physiochemical properties
Four genes proteins were showed acidic in nature (pI<7>7) with isoelectric point (pI=8.29) and molecular weight (Mw=41975.60), while SSTR2 protein showed was highly basic in nature than other selected genes with isoelectric point (pI=6.81) and molecular weight (Mw=78786.58) in human (Table 1).
Chromosomal mapping analysis
Chromosome mapping revealed that liver inflammation related genes were randomly distributed on chromosomes in human (Homo sapiens). The results showed that chromosome#2, 7 and 22 consisted only one gene. ITGA2 gene (Gene position on chromosome=150.574558), IL6 (22.727200), and MAPK1 (21.759657), located on ch2, chr7, and chr22 respectively. While chromosome chr12 (CDK2= 55.966830, and OAS2 112.978519) consisted two genes (Figure 1).

Figure 1: Chromosomal mapping.
Gene structure analysis
Web sci-pio server used as genes structure identification, there were five genes structures analyzed in human (H. sapiens). Red segment showed the gap region while red thin line indicated mismatch and blue line for sequence shift. In human, there was no mismatch or gape region found in all selected genes. ITGA2 genes (Exons: 28, Introns: 12) had greater the number of exons as compared to other genes, while IL6 gene showed only four exons with three introns. Additionally, all five genes associated with liver inflammation related genes with exon and intron showed in Figure 2.

Figure 2: Gene structures.
Motifs analysis
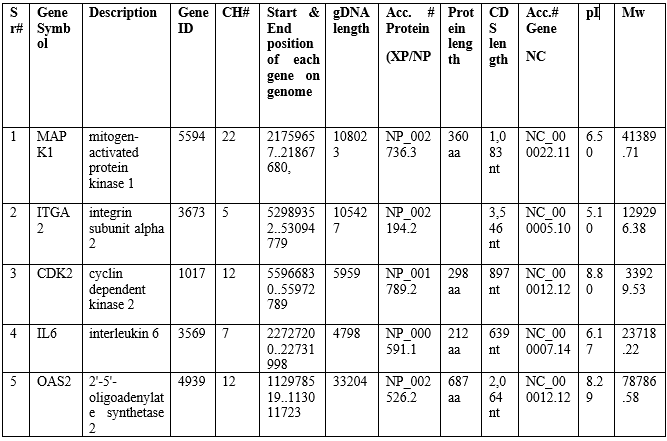
This analysis depicted the motif location, motif consensus and graphical representation of motif. Conserved motifs are involved in liver inflammation in human. Top 5 motifs were predicted; all have same frequency #2 (number of sites contributing to the construction of the motif). Motif 1 was most prominent out of top 5 motifs. Motif analysis revealed that top 5 motif located on three genes. In human, motif-1 was located OAS2 genes had width (amino acids residue) 47 with E-value = 2.2e-010. Motif-2 was located on two genes (MAPK1 and CDK2) had width 50 with E-value = 1.0e-008. Motif-3 was located on OAS2 gene had width 36 with E-value = 2.8e-005. Motif-4 was located on two genes (MAPK1 and CDK2) had width 48 with E-value = 2.3e-003. Motif-5 was located on OAS2 gene had width 21 with E-value = 2.8e-002 (Figure 3).
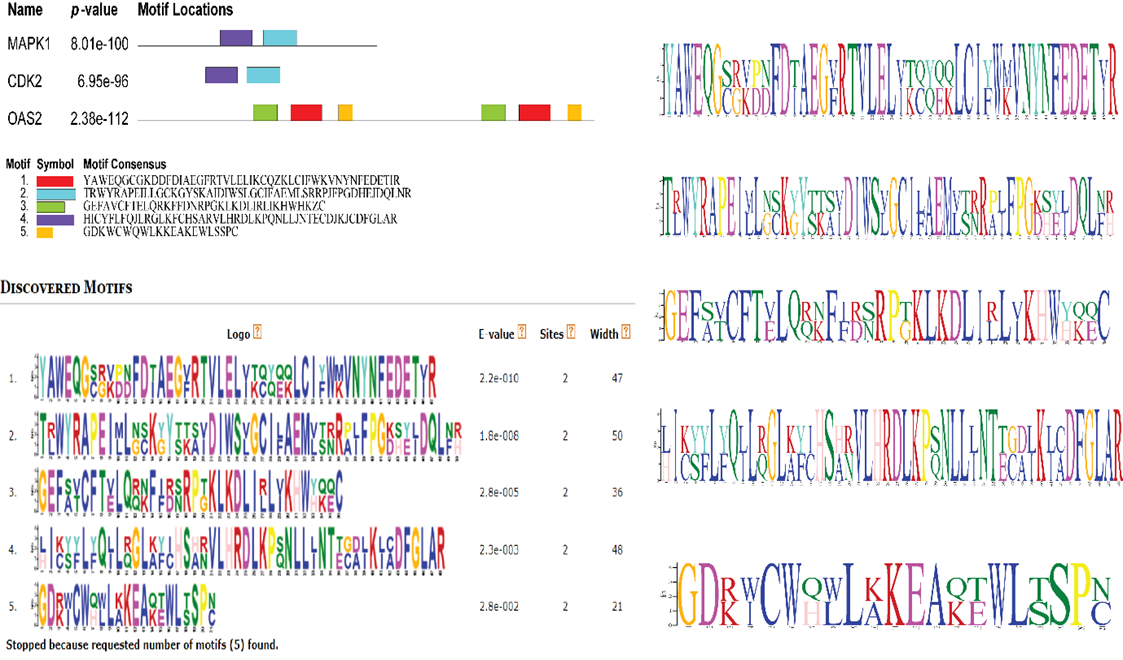
Figure 3: Motif analysis showing the motif location, motif consensus, gene frequency motif logo in Human performed by using MEME.
Protein-protein interaction
Protein control biological function solely or by association with other proteins. IL6 gene visualized by Cytoscape, revealed the protein-protein interaction. ITGA2 showed first shell of interaction with ten predicted functional partners that were ITGB1, ITGB3, FN1, ITGB2, GP6, ITGB6, COL3A1, COL1A1, COL1A2, and ITGB5 genes (Filled nods) with predicted 3D structure in human (Figure). ITGA2 gene had high degree of interaction with all prediction partners except MUC1and ADPGK gene with bitscore value 2343.9 than other genes (STRING). Empty nodes indicated unpredicted 3D structure while filled structure indicated predicted 3D structure available at PDB (data-base). Thickness of line showed degree of interaction of one gene with others (Figure 4).
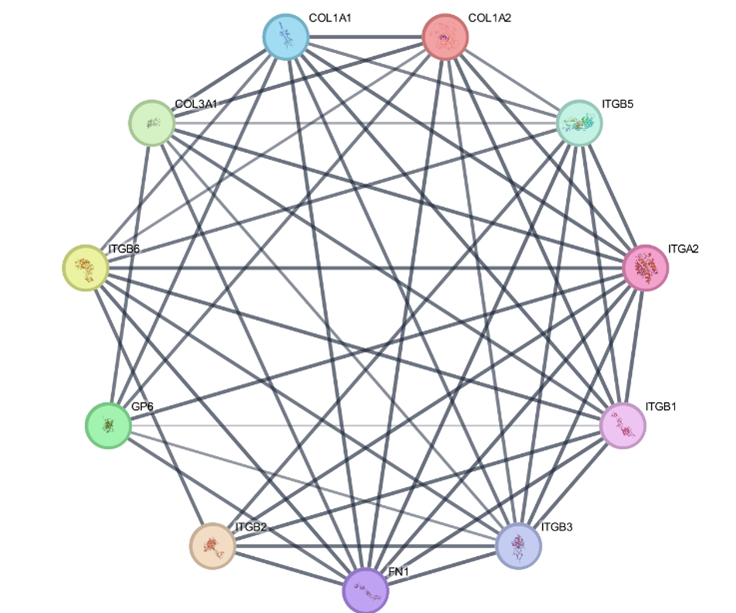
Figure 4: Protein-Protein interaction of Human visualized via Cytoscape.
Integrin alpha-2 (Alpha-2/beta-1) is the receptor for the protein laminin the protein collagen, collagen C-propeptides, fibronectin, and E-cadherin. It identifies the proline-hydroxylated motif G-F-P-G-E-R in protein. It is in charge of platelet and other cell attachment to collagen peptides regulating collagen and collagenase enzyme transcription, generating force, and organising freshly combined extracellular matrix. (Microbial Infection) Integrin ITGA2:ITGB1 serves as a binding protein on human echoviruses 1 and 8.
Gene ontology
Functional characteristics associated with proteins were determined by Gene Ontology annotation analysis. Gene Ontology results revealed liver inflammation genes annotation. The result revealed that two genes (IL6, MAPK1) involved in GO term for Endoplasmic reticulum lumen followed by cellular compartment with PV value=0.058246. There were three genes (OAS2, CDK2, MAPK1) involved in ATP binding followed by Molecular Function with PV value=0.034271. While two genes (IL6, OAS2) in defense response to virus followed by biological process with PV value=0.047379. Gene annotation discrepancy in H. sapiens (Human) showed in Table 2.
Table 2. Gene annotation discrepancy in Human.
DISCUSSION
In recent years, several researches have concentrated on the mechanism of HBV-related liver inflammation to HCC (Li et al., 2019; Ligat et al., 2019; Liu et al., 2018). However, the process by which liver inflammation develops has seldom been studied. Current research has primarily focused on one or two distinct genes, with no systematic recognising varied expressions between different levels of inflammation. This is the first study to use bioinformatics tools to analyze the gene mapping, gene, structure analysis, motif analysis except Gene ontology, Protein protein interaction. KEEG pathways and genes were identified in the development of liver inflammation. Zhao et al. (2022) conducted GO and KEGG enrichment analyses to further identify critical activities and pathways that may play a vital role in more effective antimicrobial properties in tissues of the liver with higher inflammatory grades. The pathway analysis using KEGG demonstrated that the elevated DEGs were most prevalent in Graft-versus-host disease, rheumatic arthritis, a parasitic infection the influenza A, type-1 mellitus diabetes, haematopoietic cell lineage, allograft disapproval, naturally occurring killer caused by cells cytotoxicity, and and the cytosolic DNA-sensing route (Zhao et al., 2022). My findings were comparable to previous ones, with three genes (IL6, OAS2, and MAPK1) out of five identified genes related with Influenza A. EP300, PCNA, CDKN2A, CDK2, MTOR, IL6, ITGA2, OAS2, TGFB1, and MAPK1, were pivot genes with the highest centricity and may play key roles in hepatic inflammation development. Furthermore, these genes might be potential indicators of inflammatory progression (Zhao et al., 2022). The findings would provide light on the mechanism of swelling progress, perhaps providing a hypothetical foundation and investigative indicators for liver soreness. Our current work has drawbacks, including the lack of experimental
CONCLUTION
The current study revealed that selected genes are randomly distributed across various chromosomes and are implicated in essential biological processes related to liver inflammation, there were 5 liver inflammation associated genes used in human for computational analysis. ITGA2 emerged as a central gene with a high degree of interaction, indicating its potential importance in liver inflammation networks. Gene ontology showed that most specific genes IL6, OAS2 potentially involved in defense response to virus in human. Further studies can be conducted to confirm these results and would be beneficial in screening of liver inflammation associated genes of human.
CONFLICT OF INTEREST
The authors declare no competing interests.
REFERENCES
- Cao, Y., Jia, H., Xing, M., Jin, R., Grierson, D., Gao, Z., ... & Li, X. (2021). Genome-wide analysis of MYB gene family in Chinese bayberry (Morella rubra) and identification of members regulating flavonoid biosynthesis. Frontiers in plant science, 12, 1244.
- Cobleigh MA, Robek MD. Protective and pathological properties of IL-22 in liver disease: implications for viral hepatitis. Am J Pathol. 2013; 182:21–8.
- Gao W, Fan YC, Zhang JY, et al. Emerging role of interleukin 22 in hepatitis B virus infection: a double-edged sword. J Clin Transl Hepatol. 2013; 1:103–8.
- García-Campos, M. A., Espinal-Enríquez, J., and Hernández-Lemus, E. (2015). Pathway analysis: state of the Art. Front. Physiol. 6:383. doi: 10.3389/fphys.2015. 00383
- Gregor, M. F., and Hotamisligil, G. S. (2011). Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 29, 415–445. doi: 10.1146/annurev-immunol-031210- 101322
- Hotamisligil, G. S. (2006). Inflammation and metabolic disorders. Nature 444, 860–867. doi: 10.1038/nature05485
- Hotamisligil, G. S. (2017). Inflammation, metaflammation and immunometabolic disorders. Nature 542, 177–185. doi: 10.1038/nature21363
- Hotamisligil, G. S., and Erbay, E. (2008). Nutrient sensing and inflammation in metabolic diseases. Nat. Rev. Immunol. 8, 923–934. doi: 10.1038/nri2449
- Hou XJ, Ye F, Li XY, et al. Immune response involved in liver damage and the activation of hepatic progenitor cells during liver tumorigenesis. Cell Immunol. 2018;326:52–9.
- Huang, T., Nazir, B., Altaf, R., Zang, B., Zafar, H., Paiva-Santos, A. C., ... & Ilyas, U. (2022). A meta-analysis of genome-wide gene expression differences identifies promising targets for type 2 diabetes mellitus. Frontiers in Endocrinology, 13, 985857.
- Jiao, X., Sherman, B. T., Huang, D. W., Stephens, R., Baseler, M. W., Lane, H. C., & Lempicki, R. A. (2012). DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics, 28(13), 1805-1806
- Kelesidis, I., Kelesidis, T., and Mantzoros, C. S. (2006). Adiponectin and cancer: a systematic review. Br. J. Cancer 94, 1221–1225. doi: 10.1038/sj.bjc.6603051
- Khatri, P., Sirota, M., and Butte, A. J. (2012). Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol. 8:e1002375. doi: 10.1371/journal.pcbi.1002375
- Kim SY, Kyaw YY, Cheong J. Functional interaction of endoplasmic reticulum stress and hepatitis B virus in the pathogenesis of liver diseases. World J Gastroenterol. 2017;23:7657–65.
- Kim, H., Lee, J. H., & Na, S. H. (2017, September). Predictor-estimator using multilevel task learning with stack propagation for neural quality estimation. In Proceedings of the Second Conference on Machine Translation (pp. 562-568).
- Li X, Liu X, Tian L, et al. Cytokine-mediated immunopathogenesis of hepatitis B virus infections. Clin Rev Allergy Immunol. 2016;50:41–54.
- Li ZL, Yan WT, Zhang J, et al. Identification of actual 10-year survival after hepatectomy of hbv-related hepatocellular carcinoma: a multicenter study. J Gastrointest Surg. 2019;23:288–96.
- Li, S., Shin, H. J., Ding, E. L., and van Dam, R. M. (2009). Adiponectin levels and risk of type 2 Diabetes. JAMA 302, 179–188. doi: 10.1001/jama.2009.976
- Ligat G, Schuster C, Baumert TF. HBV core variants, liver fibrosis and hepatocellular carcinoma. Hepatology. 2019;69:5–8.
- Liu F, Zeng G, Zhou S, et al. Blocking Tim-3 or/and PD-1 reverses dysfunction of tumor-infiltrating lymphocytes in HBV-related hepatocellular carcinoma. Bull Cancer. 2018;105:493–501.
- Mansouri A, Gattolliat CH, Asselah T. Mitochondrial dysfunction and signaling in chronic liver diseases. Gastroenterology. 2018;155:629–47.
- Mcdermott, J. E., Wang, J., Mitchell, H., Hafen, R., Ramey, J., and Rodland, K. D. (2013). Challenges in biomarker discovery. Expert Opin. Med. Diagn. 7, 37–51. doi: 10.1517/17530059.2012.718329.Challenges
- Minihane, A. M., Vinoy, S., Russell, W. R., Baka, A., Roche, H. M., Tuohy, K. M., et al. (2015). Low-grade inflammation, diet composition and health: current research evidence and its translation. Br. J. Nutr. 114, 999–1012. doi: 10.1017/ S0007114515002093
- Odronitz, F., Pillmann, H., Keller, O., Waack, S., & Kollmar, M. (2008). WebScipio: an online tool for the determination of gene structures using protein sequences. BMC genomics, 9(1), 1-13.
- Pradhan, A. D. (2001). C-Reactive Protein, Interleukin 6, and risk of developing type 2 Diabetes Mellitus. JAMA 286, 327–334. doi: 10.1001/jama.286. 3.327
- Shibata, R., Ouchi, N., and Murohara, T. (2009). Adiponectin and cardiovascular disease. Circ. J. 73, 608–614. doi: 10.1253/circj.CJ-09-0057
- Stefan, N., Birkenfeld, A. L., and Schulze, M. B. (2021). Global pandemics interconnected — obesity, impaired metabolic health and COVID-19. Nat. Rev. Endocrinol. 17, 135–149. doi: 10.1038/s41574-020-00462-1
- Thorand, B., Kolb, H., Baumert, J., Koenig, W., Chambless, L., Meisinger, C., et al. (2005). Elevated Levels of Interleukin-18 Predict the Development of Type 2 diabetes: results from the MONICA/KORA Augsburg study, 1984-2002. Diabetes 54, 2932–2938. doi: 10.2337/diabetes.54.10.2932
- Trehanpati N, Vyas AK. Immune regulation by t regulatory cells in hepatitis B virus-related inflammation and cancer. Scand J Immunol. 2017;85:175–81.
- Vafaee, F., Diakos, C., Kirschner, M. B., Reid, G., Michael, M. Z., Horvath, L. G., et al. (2018). A data-driven, knowledge-based approach to biomarker discovery: application to circulating microRNA markers of colorectal cancer prognosis. NPJ Syst. Biol. Appl. 4:20. doi: 10.1038/s41540-018-0056-1
- Voorrips, R. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of heredity, 93(1), 77-78.
- Yang R, Xu Y, Dai Z, et al. The immunologic role of gut microbiota in patients with chronic HBV infection. J Immunol Res. 2018; 2018:2361963.
- Zhao, J. Y., Zhong, Z. Z., Zhao, L. Y., & Li, W. (2022). Key pathways and genes in hepatitis B virus-related liver inflammation: Expression profiling and bioinformatics analysis. Medicine, 101(34), e30229.
- Zhu H, Wu J, Shen X. Genome-wide association study: new genetic insights into HBV/HCV-related hepatocellular carcinoma genomes. Scand J Gastroenterol. 2017; 52:209–15.


 Rizwan Shaukat*
Rizwan Shaukat*
 Hafsa Arshad
Hafsa Arshad



 10.5281/zenodo.13293711
10.5281/zenodo.13293711