Abstract
This review focuses on the pharmaceutical analytical profile of Remdesivir, a crucial antiviral drug used to treat viral infections like SARS-CoV-2. It emphasizes the drug's pharmacological importance and structural attributes, highlighting the critical need for precise analytical techniques to ensure its efficacy, stability, and safety. The review explores various methods for analyzing Remdesivir, each tailored to address specific aspects of its quality and performance. Non-aqueous titration assesses the drug’s solubility and stability in non-water solvents, while isothermal titration calorimetry (ITC) provides valuable insights into its interactions and thermodynamic properties. Electrochemical analysis offers sensitive and cost-effective evaluations of purity and stability. High-Performance Liquid Chromatography (HPLC) is noted for its reliability in routine quantification, with advanced techniques like LC-MS and UPLC-MS/MS ensuring the accurate detection of trace impurities and degradation products. Furthermore, spectrophotometric methods, including UV-Visible spectrophotometry and spectrofluorimetry, are highlighted for their simplicity and effectiveness in structural verification and quantitative analysis. Together, these techniques ensure Remdesivir's quality, safety, and therapeutic efficacy, supporting its critical role in pharmaceutical research and regulatory compliance.
Keywords
Pharmaceutical Analytical Methods, HPLC.
Introduction
The novel coronavirus which is from Wuhan is now causing a consideration in the medical field as the virus has become a contagious disease. The virus is been identified in late December 2019 [1]. Increasing evidence suggests that COVID-19 is transmitted between individuals [2].The number of cases kept increasing quickly, with studies forecasting that the outbreak would double in size every 1.8 days [3]. On December 31, 2019, Wuhan, located in Hubei province, China, reported multiple cases of pneumonia [4]. On January 7, 2020, Chinese health officials identified the virus as a coronavirus, showing over 95% similarity to bat coronaviruses and more than 70% similarity to SARS-CoV-2 [4,5]. The sharp rise in cases, including some unrelated to the live animal market, indicates that the disease may be transmitted through person-to-person contact [6]. In Spanish, "corona" translates to "crown " [7].COVID-19 is another term used for the coronavirus [8]. Coronaviruses (CoVs) are single-stranded, positive-sense RNA viruses that cause illness in both humans and animals [9] . Symptoms can include coughing, fever, sneezing, sore throat, headache, fatigue, chills, body aches, and a loss of taste and smell [10,11] . Symptoms are known to appear following an incubation period of 2 to 14 days [12]. The diagnosis of coronaviruses is done using reverse transcription-polymerase chain reaction (RT-PCR) and CRISPR [13] . Some antiviral drugs used in the treatment of coronavirus include remdesivir, favipiravir, lopinavir, hydroxychloroquine, nitazoxanide, and ivermectin [ 14,15,16,17,18 ]. Chemically, it is 2-Ethylbutyl (2S)-2-[[(2R, 3S, 4R, 5R)-5-[[(4-aminopyrrolo[2,1-f]-1,2,4-triazin-7-yl)-(5-cyano-3,4-dihydroxyoxolan-2-yl)-methoxy-phenoxyphosphoryl] amino] propanoate[19]. Remdesivir is a prodrug that imitates the nucleoside adenosine. Due to the low cell permeability of nucleosides, drug latentiation is essential. Antiviral medications that are nucleoside analogs are frequently modified into forms like monophosphate, ester, or phosphonamidite. These changes enhance cellular permeability, allowing the drug to enter the cell and cross the membrane, where it is subsequently converted into the nucleoside or nucleoside monophosphate form through biotransformation [20]. The cyano group at the 1’position of ribose, unique to remdesivir, is thought to prevent the molecule from binding to host mitrochondrial RNA. The highly nucleophilic C-C bond likely contributes to the cyano group’s increased stability during cellular activation. The phosphoramido group boosts the molecule’s lipophilicity, while the phosphorous-attached phenoxy group also enhances lipophilicity improving cellular permeability[21,22]. Remdesivir is a recently developed antiviral medication by Gilead Sciences, intended for the treatment of hepatitis [23,24,25]. Remdesivir is an antiviral medication used to treat coronavirus infections. It has a molecular weight of 602.59 g/mol and the chemical formula C27H35N608P. Remdesivir has a water solubility of 0.339 mg/L and pKa values of 10.23/0.65.
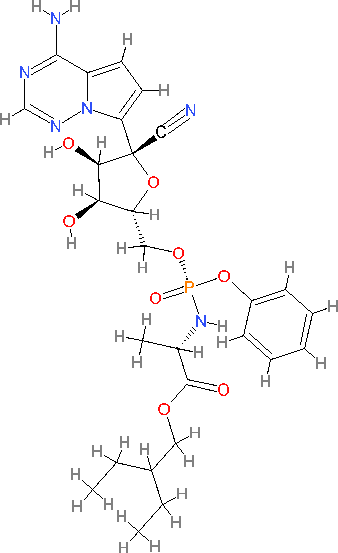
Figure 1. Structure of Remdesivir
Pharmacokinetics Properties
For the patients, remdesivir was administered intravenously (i.v.). The dosage involved 200 mg i.v. infused over 30 minutes on the first day, followed by 100 mg i.v. daily over 30 minutes from days 5 to 10. Remdesivir showed a protein binding percentage of 2% while its metabolite GS-441524 had a binding percentage of 2%.
Metabolism
Remdesivir is primarily eliminated through metabolism, glomerular filtration, and active tubular secretion (specifically for GS-441524). The drug is excreted in urine at rates of 10% for remdesivir and 49% for GS-441524, while excretion in faeces occurs at ND for remdesivir and 0.5% for GS-441524
The FDA
Pharmacodynamics Properties
Remdesivir is a broad-spectrum nucleotide prodrug that acts as an adenosine analogue[26]. And is effective against various endemic, emerging, and enzootic coronaviruses (CoV) [27,28]. These prodrugs typically exhibit greater cellular permeability and are metabolized to produce nucleosides or phosphorylated nucleosides [29,30]. Remdesivir is highly efficacious, but it has a weak solubility profile (aqeous solubility of 0.339 mg/mL, with sulfobutylether-cyclodextrin sodium (SBECD) enhancing solubility to 5 mg/mL, logP values ranging from 2.0 to 2.2, and a pKa of 10.23). Additionally, it is primarily eliminated through the kidneys (78%) [31]. On the first day, a 200 mg of loading dose is administered intravenously, after which a daily maintenance dose of 100 mg is given via a 30 minute infusion for a maximum of 10 days [32]. Host cells transform the C-adenosine analogue (GS-441524) and its monophosphoramidate prodrud, Remdesivir, into an active nucleoside triphosphate (NTP). Inside the host cells ,Remdesivir is hydrolyzed to produce and alanine metabolites (GS-704277), which is then converted into monophosphate and diphosphate derivatives before being triphosphorylated[33,24]. The nucleotide analogue Remdesivir triphosphate acts as a substrate for the viral RNA-dependent RNA polymerase (RdRp), playing a crucial role in controlling the replication of SARS-CoV-2[34]. Remdesivir also targets the viral RNA-dependent RNA polymerase, leading to the inhibition of SARS-CoV-1 in laboratory studies[35].. Research has demonstrated that using RMD treatment can help decelerate the advancement of severe respiratory illness[36]. On October 22, 2020, the FDA approved Remdesivir as a treatment for COVID-19 due to positive clinical outcomes and its temporary authorization for compassionate use[36,27]. The presence of a distinct exoribonuclease (ExoN) has posed a challenge in creating potent nucleoside analogues to combat coronaviruses. Exonuclease (ExoN) functions as a proofreading enzyme, rectifying mistakes in the RNA chain during the elongation process [34, 37]. Remdesivir demonstrates enhanced effectiveness against viruses that do not have ExoN; yet, it is capable of partially resisting proofreading and sustaining substantial antiviral efficacy even in the presence of ExoN [38]. ExoN seamlessly integrates into replicating RNA with greater efficiency compared to native nucleotides, hence diminishing remdesivir's effectiveness [34,39,40].
Polymers
At present, Remdesivir is administered via injectables, which introduces certain challenges. The function of remdesivir polymorphs are fascinating and warrant investigation to explore all possible mechanisms for its delivery[41]. Since it greatly affects product stability, tableting and compression characteristics, dissolution rate and profiles, and conclusively pharmacokinetic, the nature of therapeutics drugs-including co-crystals, salts, and polymorphism-should be taken into account in the fight against COVID-19[42,43]. Remdesivir exists in two solvent-free polymorphic forms, RDV-I and RDV-II, each exhibiting distinct physiochemical and pharmacokinetics characteristics, Although remdesivir has four polymorphs (RDV-I through RDV-IV), its exact molecular connectivity remains uncertain due to the absence of single-crystal diffraction data. RDV-III was believed to contain a harmful CH2Cl2 solvent, while RDV-IV was considered unstable in solution and quickly converted to RDV-II [44]. Available Market Formulations of Remdesivir
Remdesivir is the newest medication in the pharmaceutical industry for treating coronavirus and is n parentral form. Table 1 provides information on several companies and their respective brand names
Table 1 . Selected pharmaceutical formulations of Remdesivir available in the market
|
S.NO
|
Company
|
Brand Name
|
Available
Dosage (mg)
|
References
|
|
1.
|
Cipla
|
CIPREMI
|
100mg
|
[45]
|
|
2.
|
Hetero HC
|
COVIFOR
|
100mg
|
[46]
|
|
3.
|
Mylan
|
DESREM
|
100mg
|
[47]
|
|
4.
|
Zydus Cadila
|
REMDAC
|
100mg
|
[48]
|
Analytical methods for Quantifying Remdesivir
Non-Aqeous Titration
The WHO has proposed a draft for the inclusion of Remdesivir in the International Pharmacopoeia. They recommend a non-aqeous titration method for assaying Remdesivir, involving the dissolution of 0.4 g in 50 mL of glacial acetic acid, followed by titration with 0.1 mol/L perchloric acid. Each milliliter of 0.1 mol/L perchloric acid corresponds to 60.26 mg of Remdesivir [49].
Multiple Isothermal Titration Calorimetry (Itc)
Isothermal titration calorimetry (ITC) under multiple conditions.Experts from universities in Spain and the USA conducted various isothermal titration calorimetry (ITC) experiments in conjunction with computational molecular dynamics simulations. Their aim was to explore the structural and thermodynamic elements of the connection between CAPTISOL and either neutral or protonated Remdesivir. At a pH of 3, thorough examination of the calorimetric data revealed a robust complex with a high association constant of 10^4. The profiles of the potential mean force (PMF) indicated that lower association constant values correspond to less tightly bound structures with fewer sulfobutylether (SBE) substitutions. This finding is in agreement with the results from isothermal titration calorimetry (ITC) that emphasized how the affinity is notably impacted by both the quantity and positioning of CAPTISOL-containing structures [50].
Electrochemical Method Of Analysis
Theoretical analysis of the electrochemical determination of the anti-COVID-19 drug Remdesivir was conducted for the first time. This study explored an anodic approach using the Squaring Dye-Ag202 combination. The mechanism of the electroanalytical process is complex, suggesting a dynamic nature. A mathematical model study, grounded in linear stability theory and bifurcation analysis, demonstrates that the composite achieves genuine electroanalytical efficiency as an electrode modification, despite being a modification itself [51].
Typically both qualitative and quantitative analysis of remdesivir is conducted using methods such as HPLC, LCMS, fluorometry, and UV-Visible spectrosccopy
High-Performance Liquid Chromatography Method (Hplc)
In HPLC, each chemical substance in the sample are separated according to its particular affinity for the absorbent material in the mobile phase or in the column, enabling the different components to move at different rates and achieve separation[52]. The separation of HPLC is influenced by factors such as the polarity, pH, flow rate, and other characteristics of the mobile phase, along with the type and nature of the stationary phase. Additionally, environmental factors like temperature, as well as the type and settings of the detector, also play a role[53]. For non-pharmacopeial products, an alternative method can be improved by reducing costs and time while enhancing precision and robustness. When a proposed alternative method is intended to replace an existing procedure, comparative laboratory data outlining its advantages and limitations are typically provided. The HPLC technique is designed to separate and quantify the main active compound, reactive impurities, synthetic intermediates, and degradation products [54].
The WHO proposal monograph for Remdesivir [55] outlines an HPLC method for determining Remdesivir and its related compounds in the finished product. The method employs a stainless steel column (4.6 mm × 25 cm) packed with end-capped silica gel particles bonded with octadecylsilyl groups (5 ?m). The mobile phase consists of 35 parts phosphoric acid solution and 65 parts methanol, with a flow rate of 1.0 mL per minute. Detection is performed using a UV spectrophotometer set at a wavelength of 237 nm, with the column temperature maintained at 25 °C.
For analysis, 5 ?L of both standard and test solutions are injected, and the chromatograms are recorded over 25 minutes. The percentage content of C17H35N6O8P in the sample is determined by comparing the peak areas corresponding to Remdesivir in the standard and test solution chromatograms, using the specified content of C17H35N6O8P in Remdesivir as a reference.
Umstead [56], in the DAICEL Chiral Technologies proposal, detailed an HPLC method for the chiral separation and determination of Remdesivir and its key starting materials in the final product. The method employed a CHIRALPAK® IA-3 column (250 mm × 4.6 mm i.d.) and utilized a mobile phase comprising n-Hexane, Ethanol, IPA, Ethanolamine, and Formic Acid in a ratio of 80:5:15:0.05:0.1 (v/v/v/v/v), with a flow rate of 1.5 mL/min. Detection was performed using a UV spectrophotometer at 245 nm with a reference wavelength of 450 nm. The column temperature was maintained at 40 °C. A standard solution (1.0 mg/mL) was prepared in a mixture of Hexane and Ethanol (50/50, v/v), and 10 ?L was injected for analysis, with chromatograms recorded over 40 minutes.
Separately, the Waters proposal investigated analytical methods for Remdesivir, focusing on its separation and determination to monitor biotransformation events.
Alden et al. [57] described a liquid chromatography method combined with optical or mass spectrometric detection for analyzing nucleotide analogs using tributylamine as an ion-pairing agent. The study employed an Atlantis PREMIER BEH C18 AX 1.7 ?m column (2.1 × 50 mm) with an incremental gradient elution to separate analytes across a broad range of polarities in a single run. The mobile phases were prepared on-line using IonHance buffer concentrates diluted at a 1:5 ratio to achieve final concentrations of 100 mM in 4?etonitrile for IonHance CX-MS Concentrate A (pH 5) and 200 mM in 4?etonitrile for IonHance Ammonium Acetate (pH 6.8). The gradient transitioned from 5 mM ammonium acetate (pH 6.8) in 0?etonitrile to 20 mM ammonium acetate (pH 6.8) in 60?etonitrile within 4 minutes, followed by a 0.5-minute return to the initial concentration. A longer 8-minute gradient also showed promising results. The flow rat e was 0.5 mL/min, with the column maintained at 50 °C and samples at 12 °C. Detection was performed using an Acquity Premier PDA Detector and an Acquity QDa Mass Detector. Reddy et al. [58] developed a validated HPLC method with a UV detector to quantify degradation products in Remdesivir injectable formulations. The method utilized a Kromasil KR100-5 C18 column (250 mm × 4.5 mm, 5 ?m) and was validated according to current regulatory guidelines, including those from the International Conference on Harmonization (ICH). The method effectively measured degradation products at concentrations ranging from the quantification level to 200% of the specification for both known and unknown impurities and was successfully applied to analyze Remdesivir injectable products.
Patel et al. [59] designed an RP-HPLC technique for separating and quantifying Remdesivir in its API form. The method used a C18 column (250 mm × 4.6 mm, 5 ?m) with a mobile phase of Buffer (pH 5.0): Acetonitrile (30:70) at a flow rate of 1 mL/min and detection at 253 nm. The retention time was 4.402 minutes, and the method demonstrated acceptable linearity, accuracy, and precision, with a linearity range of 10–30 ?g/mL. Stress testing revealed significant degradation under alkaline conditions, highlighting its stability.
.Liquid Chromatography Mass Spectrometry Method
An LC-MS system consists of an autosampler, HPLC system, ionization source (which links the LC to the MS), and the mass spectrophotometer. It’s important to note that the mobile phases and flow rates compatible with HPLC-MS connections are limited, Restrictions also apply to mobile phase modifiers, which typically need to be volatile. These modifiers are chemicals added to the mobile phase to enhance the chromatography of target analytes, with common examples including ammonium acetate, acetic acid, and formic acid. Numerous studies highlight the key HPLC parameters essential for LC-MS analyses [60,61,62,]. The two most commonly used ionization sources in LC-MS are electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI); both have become standard features in mass spectrometers for LC-MS applications. ESI and APCI operate at atmospheric pressure, where a combination of heat and high voltage enables the necessary ionization to generate ions. In ESI, a high-voltage field (3-5kV) induces the nebulization of the column effluent, creating charged droplets move toward the mass analyzer’s inlet, they shrink, and individual ions are released in a process known as ‘’ion evaporation.’’ The MS system then separates these ions [63,64,].
Table 3. LC-MS method for estimation of remdesivir
|
Compound
(Remdesivir)
|
Ionization
Method
|
Capillary
temperature
|
Ionization
voltage
|
Solvent
Mixture
|
Mass Spectrophotometer used
|
Run time
|
References
|
|
Method 1
|
Heated
Electrospray
Ionization
|
270°C
|
4300 V
Spray
voltage
|
90%:10%
Acetonitrile:
Water
|
Thermo Fisher
Scientific TSO Altis triple quadrupole MS
|
1.2 min
|
[65]
|
|
Method 2
|
Electrospray
Ionization
|
150°C
|
__
|
Methanol:
Water
(1:1; v/v)
|
Triple quadrupole
MS
|
5.0 min
|
[66]
|
|
Method 3
|
Electrospray
Ionization
|
150°C
|
__
|
% formic acid (v/v) in
Acetonitrile
|
Waters Xevo ® TQ-S micro tandem quadrupole mass
spectrometer
|
4
Min
|
[67]
|
|
Method 4
|
Electrospray
Ionization
|
350°C
|
__
|
0.1% formic acid (A) andacetonitrile (B) starting from 0% of (B) to 100%
|
TSQ Endura triple
Quadrupole mass
Spectrometer (ThermoFisher)
|
5
min
|
[68]
|
|
Method 5
|
Electrospray
Ionization
|
350°C
|
3500V
|
10Mm sodium
formate buffer in 0.1% formic acid (A) and acetonitrile (B)
|
Triple quadrupole
MS
|
5
Min
|
[69]
|
|
Method 6
|
Turbo Ion Spray
|
650°C
|
4500V
|
Acetonitrile: Dimethyl Sulfoxide at
50:50 (v:v)
|
Triple quadrupole
MS
|
3.4
Min
|
[70]
|
Ultra Performance Liquid Chromatography-Tandem Mass Spectrophotometer (Uplc-Ms/Ms)
The study conducted by Du and colleagues. A method was developed by [71] using HPLC-MS/MS to measure GS-441524 (Nuc), the active metabolite of remdesivir, in rat plasma samples through a protein precipitation process. Gradient elution was utilized using mobile phase A, consisting of a mixture of 95% acetonitrile and 5% water with 0.1% formic acid is used in mobile phase A, while mobile phase B consists of a mixture of 99 parts water to 1 part ACN with 0.1% Formic acid solutions (1% concentration) were freshly prepared. For baseline separation, the gradient elution protocol was as follows: 1.2min at 99% B, followed by 5?rom 1.2-3.5 min, then back to 99?or 3.5-4.5 min. Methanol:water (1:1,v/v) was used to prevent carryover. Chromatographic separation took place on a Waters X Bridge C18 column (50 × 2.1 mm, 3.5 ?m) with a runtime of 4.5 min. In electrospray positive ion mode, the selected reaction monitoring transitions for Nuc were m/z 292.2? 163.2, while the internal standard (carbamazepine) used transitions of 237.1 ? 194.1. The column was maintained at a temperature of 40 °C. A 1.0 mg/mL solution of GS-441524 (Nuc) was prepared in MeOH:water (1:1, v/v), 1 ?L injected and the chromatogram recorded for 4.5 min. Nuc’s linearity was determined using the internal standard, and the calibration curve showed linearity over a range of 2-1000ng/mL, with a correlation coefficient (r) above 0.990
Pasupuleti et al. [72] created an analytical method for monitoring Remdesivir’s drug profile in human plasma for pharmacokinetics (PK) and therapeutic drug monitoring (TDM)). To rapidly detect Remdesivir in human plasma, they developed a vortex-assisted salt-induced liquid-liquid micro-extraction (VA-SI-LLME) method combined with UHPLC-PDA and UHPLC- MS/MS. This method involves a single protein precipitation step with hydrochloric acid followed by extraction with acetonitrile for analysis. Optimal conditions include 500 µL of acetonitrile and 2.5 g of ammonium sulfate with a 2-minute vortex extraction. Correlation coefficients of 0.9969 for UHPLC-PDA ( measured at 254 nm ) and 0.9990 for UHPLC-MS/MS ( with positive ion electrospray ionization and transitions of m/z 603.1 ? m/z 402.20 and m/z 603.1 ?m/z 199.90) were achieved. Detection and quantification limits were 1.5 and 5ng/Ml for UHPLC-PDA, and 0.3 and 1 ng/ml for UHPLC-MS/MS, respectively. The method provided extraction recoveries between 90.79-116.74% for UHPLC-PDA and 85.68-101.34% for UHPLC-MS/MS, with intraday and interday precision within 9.59% for both techniques.
Avataneo and colleagues. Researchers at reference [73] have created a highly efficient UHPLC-MS/MS technique for quantifying Remdesivir and GS-441524 levels in plasma samples obtained from healthy donors, employing protein precipitation during sample preparation. A chromatographic separation was carried out using an Acquity HSS T3 column, employing a gradient of water and acetonitrile with a concentration of 0. A solution of 0. 05% formic acid. The validation process adhered to the guidelines set by EMA and FDA, resulting in calibration curves showing linearity through zero with r?2; values exceeding 0. 998 shows a high level of precision. A weighting factor of 1/x was implemented to enhance precision when dealing with lower concentrations
.Alvarez and colleagues. A method was introduced by [74] that utilized LC coupled with triple quadrupole mass spectrometry for the identification of Remdesivir and GS-441524 in plasma. Following a straightforward protein precipitation with methanol, the analytes were selectively separated on a Kinetex Polar C18 column, utilizing electrospray ionization in positive mode. Calibration demonstrated linearity between 1 and 5000 µg/L for Remdesivir, and between 5 and 2500 µg/L for GS-441524, with detection limits of 0. Five and two micrograms per liter, respectively. The method demonstrated accuracy levels under 14. An accuracy level of 89% is needed, along with a 7?ctor. Six to one hundred ten. Remdesivir displayed increased stability in NaF plasma, exhibiting swift initial drops that were succeeded by gradual reductions in metabolite concentrations.
Hu et al. [75] developed an HPLC-MS/MS method to separate the active metabolite Remdesivir triphosphate (RTP) and its precursor Remdesivir monophosphate (RMP) using a BioBasic AX column. By optimizing retention with an anion exchange column and enhancing matrix stability, they achieved quantification limits of 20 nM for RMP and 10 nM for RTP. Validation showed precision (RSD) of 11.9% for RMP and 11.4% for RTP, with accuracy between 93.6–103% for RMP and 94.5–107% for RTP.
Xiao et al. [76] implemented an LC-MS/MS method for Remdesivir, GS-441524, and GS-704277 in human plasma, addressing stability issues with formic acid treatment and using distinct electrospray ionization (ESI) modes to maximize sensitivity. Calibration was linear across ranges of 4–4000 ng/mL for Remdesivir and 2–2000 ng/mL for GS-441524 and GS-704277. Precision was below 6.6%, and accuracy was within 11.5%, with analyte stability confirmed for extended frozen storage. Reckers et al. [77] established an LC-MS/MS method for Remdesivir, GS-441524, and dexamethasone, applied to blood samples from COVID-19 patients. Detection limits were 0.0375 ng/mL for Remdesivir, 0.375 ng/mL for GS-441524, and 3.75 ng/mL for dexamethasone. They noted modest within-patient variability but significant variability across patients, highlighting the need for therapeutic drug monitoring and dose adjustments in COVID-19 treatment.
Spectroscopic Method Of Analysis
Spectrophotometry
Bulduk and Akbel developed and validated UV spectrophotometric techniques for measuring Remdesivir in pharmaceutical items. UV spectra were acquired across a wavelength range of 200-800 nm using deionized water as the solvent, with a selected wavelength of 247 nm. The methods have been validated in accordance with the guidelines of ICHQ2 (R1), demonstrating outstanding outcomes in terms of linearity, accuracy, and recovery. Correlation coefficients exceeded 0. The concentration of 999 falls within the range of 10-60mg mL-1 [78].
Spectrofluorometric Method
Heba Elmansi et al. [79] noted considerable interest in fluorescene spectroscopy deu to its strong analytical performance and environmental sustainability. For pharmaceutical quality control, methods need to be sensitive, efficient, and cost-effective to achieve high throughput at a reasonable cost. The study on Remdesivir relied on spontaneous fluorescence measurements at pH 4, within wavelengths 244-405nm. Calibration was performed across a 1.0-65.0 ng/mL range, with various factors examined to maximize sensitivity, yielding detection and quantification limits of 0.287 and 0.871ng/mL, respectively. This is thought to be the first spectrofluorimetric method developed for quantifying Remdesivir. The method was also applied to detect the drug in prepared IV infusions and spiked huma plasma samples. Statistical analyses confirmed the accuracy and reliability of the method. The environmental friendliness of the developed approach was highlighted using the Green Analytical Procedure Index (GAPI) and the AGREE- analytical greenness meter, both recently developed tools for assessing environmental impact. According to Tamer Z. Attia at el., Remdesivir, a treatment for COVID-19 was analyzed in vials and in samples spiked with human plasma using spectrofluorimetric methods [80]. A sensitive, straightforward, and eco-friendly spectrofluorimetric method has been developed for quantifying Remdesivir (REM) in pharmaceutical formulations and in spiked human plasma. This method relies on detecting REM’s natural fluorescence in distilled water, with emission at 410nm and excitation at 241nm, yielding a linear response over a concentration range of 50.00 to 500.00ng/mL. Sensitivity for REM was further enhanced by micellar formation using 2.00% w/v sodium dodecyl sulfate (SDS), resulting in a linear range from 10.00 to 350.00 ng/mL, along with detection and quantification limits of 2.34 ng/mL and 7.10 ng/mL, respectively. Key analytical parameters were meticulously examined. Following FDA and International Council for Harmonization (ICH) guidelines, the validation study confirmed the method’s reliability.
Uv- Visible Spectrophotometry Method
The UV-visible spectrophotometry equipment is commonly used, providing analysts with substantial benefits because of its efficiency, cost-effectiveness, and easy scalability[81]. UV spectrophotometric and HPLC methods have been created and verified for quantitatively assessing Remdesivir in pharmaceutical formulations. An HPLC analysis was performed on a C-18 column with a mobile phase consisting of a 20 mM KH2PO4 solution and acetonitrile (50:50, v/v) running at a flow rate of 12mL/min. UV spectra ranging from 200 to 800 nm were obtained using de-ionized water as the solvent, with a detection wavelength of 247 nm. The validation of these methods was carried out in accordance with the ICH guideline Q2 (R1). Both methods demonstrated outstanding linearity, accuracy, and recovery, without any interference from non-medicinal substances in terms of spectral or chromatographic analyses. The correlation coefficients have surpassed 0. 999 demonstrated efficacy across a concentration range of 10-60mg/mL [82].
CONCLUSION
The review paper effectively highlights the overall advancements in methods for assessing drugs. The authors have conducted a detailed analysis of Remdesivir, a prodrug predominantly utilized as a primary treatment option for SARS-CoV-1. Their goal is to offer a comprehensive, comparative, and effective analytical overview of this medication. The study delves into the current trends in analytical techniques used to detect Remdesivir, showcasing their advantageous influence on pharmaceutical analysis. The paper explores pharmacodynamics and pharmacokinetics, delving into different analytical methods utilized in studying Remdesivir such as HPLC, LCMS, UV-Visible spectroscopy, and fluorescence techniques. Concurrent bioactivity assessments can provide valuable insights to assist in the continuous process of drug discovery and evaluation. Further research on Remdesivir is still needed. It is crucial to perform both qualitative and quantitative determinations to guarantee the safety and effectiveness of medications in different matrices. Until now, there has not been any stability-indicating method disclosed for quantifying impurities and degradation products of remdesivir in pharmaceutical formulations. Hence, it is crucial to conduct forced degradation studies in compliance with the guidelines of the International Conference on Harmonization and to establish a specific and validated HPLC stability-indicating technique. These studies offer reliable and validated analytical techniques to track Remdesivir and its metabolites, thereby endorsing their clinical application and guaranteeing precise and consistent therapeutic drug monitoring in patient samples.
REFERENCES
- Rothe, C., Schunk, M., Sothman, P., Bretzel, G., Froeschl, G., Wallrauch, C., Zimmer, T., Theil, V., Janke, C., Guggemos, W., Seilmaier, M., Drosten, C., Vollmar, P., Zwirglmaier, K., Zange, R., Hoelscher, M. N. Engl. J. Med., 2020, vol. 382(10), pp.970-971. Doi:10.1056/nejmc2001468.
- Chan, J . F .-W .,Yuan, S., Kok, K.-H., To, K. K.-W., Chu, H., Yang, J., Xing, F ., Liu, J ., Yip, C. C.-Y ., Poon,R. W .-S., Tsoi, H.-W., Lo, S. K.-F., Chan, K.-H., Poon, V. K.-M., Chan, W.-M., Ip, J.D., Cai, J.-P., Cheng, V . C.-C., Chen, H., Hui, C., Yuen, K.-Y . Lancet, 2020, vol. 395(10223), pp,514-523. Doi: 10.1016/S0140-6736(20)30154-9.
- Li, Q., Guan,, XX., Wu, P., Wang, X., Zhou, L., Tong, Y., Ren, Leung, K., Lau, E., Wong, J., King, X., Xing, X., Xiang, N., Wu, y ., Li, C., Chen, Q., Li, D., Liu, T., Zhao, J., Liu, M., Tu, W., Chen,C., Jin, L., Yang, R., Wang, Q., Zhao, S., Wang, R., Liu, H., Luo, Y., Liu, Y., Shao, G., Li, Y., Shao, G., LI, h., Tao, Z., Yang, Y., Deng, Z., Liu, B., Ma, Z., Zhang, Y., Shi, G., Lam, T., Wu, J., Gao, G., Cowling, B., Yang, B., Leung, G., Feng, Z. N. Engl. J, Med. 2020, vol.382(13), pp.1199-1207 . doi: 10.1056/nejmoa2001316.
- Chang, F. Y., Chen, H. C., Chen, P. J., Ho, M. S., Hsieh, S. l., Lin, J. C., Liu, F. T., Sytwu, H. K. J. Biomed. Sci., 2020, vol. 27(1), pp- 1-13. Doi: 10.1186/s2929-020-0063-w.
- Singhal, T. Indian J. Pedatr, 2020, vol. 87(4), pp. 281-286. doi: 10.1007/s12098-020-032636.
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., Liu, M., Xia, Y., Gao, H., Guo, L., Xie, J., Wang, G., Jiang, R., Gao, Z., Jin, Q., Wang, J., Cao, B. . Lancet.,2020, vol. 395(10223), pp. 497-506. Doi: 10.1007/s12098-020-03263-6.
- Alinia-ahandani, E., Milad, s. Journal of Medical and Biological Science Research. 2020, vol. 6 (2). Doi: 10.36630/jmbsr.
- Ing, E. B., Xu, Q. A., Salimi, A., & Torun, N. Occup. Med. 2020, vol. 70(5), pp. 370-374. Doi: 10.1093/occmed/kqaaa088.
- Habas, K., Nganwuchu, C., Shahzad, F., Gopalan, R., Haque, M., Rahman, S., Majumder, A, A., & Nasim, T Expert Rev. Anti. Infect. Ther, 2020, vol. 18(120, pp- 1201-1211. Doi: 10.1080/14787210.2020.1797487.
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., Tan, W. N. Engl, J. Med, 2020, vol. 382(8), pp- 727 -733. Doi: 10.1056/nejmoa2001017.
- Singh, R., Sarsaiya, S., Singh, T. A., Singh, T., Pandey, L. K., Pandey, P. K. Journal of Drug Delivery and Therapeutics, 2021, vol. 11(2), pp. 118-120. doi:10.22270/jddt.v11i2-s.4644.
- Rothan, H. A., Byrareddy, S. N. J. Autoimmun, 2020, vol. 109, pp. 102433. doi: 10.1016/j.jaut.2020.102433.
- Zhang, F., Abudayyeh, O. O., Gootenberg, J. S., Sciences, C., Mathers, L. Bioarchive, 2020, vol. 20200321 pp. 1–8.
- Joshi, S., Parkar, J., Ansari, A., Vora, A., Talwar, D., Tiwaskar, M., Patil, S., Barkate, H, Int. J. Infect. Dis, 2021, vol.102, pp. 501–508. doi: 10.1016/j.ijid.2020.10.069.
- Amani, B., Khanijahani, A., Amani, B., Hashemi, P, J Pharm Pharm sci, 2021, vol. 24, pp. 246 –257
- Chen, Y., Li, M. X., Lu, G. D., Shen, H. M., Zhou, J, Int. J. Biol. Sci, 2021, vol. 17(6), pp. 1538–1546. doi: 10.7150/ijbs.59547.
- Rocco, P., Silva, P., Cruz, F., Melo. M., Tierno, P., Moura, M., de Oliveira, L., Lima, C., Dos Santos, E., Junior, W., Fernandes, A., Franchini, K., Magri, E., de Moraes, N., Gonçalves, J., Carbonieri, M., Dos Santos, I., Paes, N., Maciel, P., Rocha, R., de Carvalho, A., Alves, P., Proença-Módena, J., Cordeiro A., Trivella D., Marques R., Luiz R., Pelosi P., e Silva J, Eur. Respir. J, 2021, vol. 58(1). doi:10.1183/13993003.03725-2020
- Pandey, S., Pathak, S. K., Pandey, A., Salunke, A. A., Chawla, J., Sharma, A., Sharma,S., Thivari, P., Ratna, H. V. K, Diabetes Metab. Syndr. Clin. Res. Rev, 2020, vol. 14(6), pp.1921–1922. doi:10.1183/13993003.03725-20
- B., V., BABU, A., MARIA, F., ANTONY, S, Int. J. Curr. Pharm. Res, 2020, vol. 12(6), pp. 20–23. doi: 10.22159/ijcpr.2020v12i6.40298.
- Al-tannak, N. F., Novotny, L, Sci. Pharm, 2020, vol. 88(29), pp. 1-12. doi:10.3390/scipharm88020029
- Pruijssers, A. J., Denison, M. R, Curr. Opin. Virol, 2019, vol. 35, pp. 57–62. doi: 10.1016/j.coviro.2019.04.002.
- Kadioglu, O., Saeed, M., Greten, H. J., Efferth, T, Comput. Biol. Med, 2021, vol. 133, Pages-104359. doi: 10.1016/j.compbiomed.2021.104359
- Chatterjee, S, Drug Res, 2021, vol. 71(3), pp.138–148. doi: 10.1055/a-1288-4078.
- Warren, T. K., Jordan, R., Lo, M. K., Ray, A. S., Mackman, R. L., Soloveva, V., Siegel, D., Perron, M., Bannister, R., Hui, H. C., Larson, N., Strickley, R., Wells, J., Stuthman, K. S., Van Tongeren, S. A., Garza, N. L., Donnelly, G., Shurtleff, A. C., Retterer, C. J., GharaibehD., Zamani R., Kenny, T., Eaton, B., Grimes, E., Welch, L., Gomba, L., Wilhelmsen, C., Nichols, D., Nuss J., Nagle, E., Kugelman, J., Palacios, G., Doerffler, E., Neville, S., Carra E., Clarke, M., Zhang, L., Lew, W., Ross, B., Wang, Q., Chun, K., Wolfe, L., Babusis, D., Park Y., Stray, K., Trancheva, I., Feng J., Barauskas O., Xu, Y., Wong, P., Braun, M., Flint, M., McMullan, L., Chen, S., Fearns, R., Swaminathan, S., Mayers, D., Spiropoulou, C., Lee, W., Nichol, S., Cihlar, T., Bavari, S. Nature, 2016, vol. 531(7594), pp. 381–385. doi: 10.1038/nature17180.
- Mulangu, S., Dodd, L. E., Davey, R. T., Tshiani Mbaya, O., Proschan, M., Mukadi, D., Lusakibanza Manzo, M., Nzolo, D., Tshomba Oloma, A., Ibanda, A., Ali, R., Coulibaly, S., Levine, A. C., Grais, R., Diaz, J., Lane, H. C., Muyembe-Tamfum, J.-J., the PALM Writing Group. N. Engl. J. Med, 2019, vol. 381(24), pp. 2293–2303. doi:10.1056/nejmoa1910993.
- Ahmed, A. B., Gamal, M., Naguib, I. A., Ali, H. M., & Abdallah, F. F. Microchem. J, 2022, vol. 176, pp.107242. doi: 10.1016/j.microc.2022.107242.
- Grein, J., Ohmagari, N., Shin, D., Diaz, G., Asperges, E., Castagna, A., Feldt, T., Green, G., Green, M. L., Lescure, F.-X., Nicastri, E., Oda, R., Yo, K., Quiros-Roldan, E., Studemeister, A., Redinski, J., Ahmed, S., Bernett, J., Chelliah, D., Chen, D., Chihara, S., Cohen, S., Cunningham, J., D’Arminio Monforte, A., Ismail, S., Kato, H., Lapadula. G., L’Her, E., Maeno, T., Majumder, S., Massari, M., Mora-Rillo, M., Mutoh, Y., Nguyen, D., Verweij, E., Zoufaly, A., Osinusi, A., DeZure. A., Zhao, Y., Zhong, L., Chokkalingam, A., Elboudwarej, E., Telep, L., Timbs, L., Henne, I., Sellers, S., Cao, H., Tan, S., Winterbourne, L., Desai, P., Mera, R., Gaggar, A., Myers, R., Brainard, D., Childs, R, N. Engl. J. Med, 2020, vol. 382(24), pp. 2327–2336. doi:10.1056/nejmoa2007016.
- Sheahan, T. P., Sims, A. C., Graham, R. L., Menachery, V. D., Gralinski, L. E., Case, J. B., Leist, S. R., Pyrc, K., Feng, J.Y., Trantcheva, I., Bannister, R., Park, Y., Babusis, D., Clarke, M. O., MacKman, R. L., Spahn, J. E., Palmiotti, C. A., Siegel, D., Ray, A., Cihlar, T., Jordan, R., Denison, M, Sci. Transl. Med, 2017, vol. 9(396), pp.1–11. doi:10.1126/scitranslmed.aal3653.
- De Clercq, E, Nat. Rev. Drug Discov, 2002, vol. 1(1), pp. 13–25. doi: 10.1038/nrd703.
- Seley-Radtke, K. L., Yates, M. K. Antiviral Res, 2018, vol. 154, pp. 66–86. doi:10.1016/j.antiviral.2018.04.004.
- Hanafin, P. O., Jermain, B., Hickey, A. J., Kabanov, A. V., Kashuba, A. D. M., Sheahan, T. P., Rao, G. G, CPT PharmacometricsSyst. Pharmacol, 2021, vol. 10(2), pp. 89–99. doi: 10.1002/psp4.12584.
- Spinner, C. D., Gottlieb, R. L., Criner, G. J., Arribas López, J. R., Cattelan, A. M., Soriano Viladomiu, A., Ogbuagu, O., Malhotra, P., Mullane, K. M., Castagna, A., Chai, L. Y. A., Roestenberg, M., Tsang, O. T. Y., Bernasconi, E., Le Turnier,P., Chang, S. C., Sengupta, D., Hyland, R. H., Osinusi, A. O., Cao, H., Blair, C., Wang, H., Gaggar, A., Brainard, D., McPhail, M., Bhagani, S., Ahn, M., Sanyal, A., Huhn, G., Marty, F. M, JAMA -J. Am. Med. Assoc, 2020, vol. 324(11), pp.1048–1057. doi:10.1001/jama.2020.16349.
- Sun, D. AAPS J, 2020, vol. 22(4), pp.77. doi:10.1208/s12248-020-00459-8.
- Gordon, C. J., Tchesnokov, E. P., Woolner, E., Perry, J. K., Feng, J. Y., Porter, D. P., Gotte, M., J. Biol. Chem, 2020, vol. 295(20), Pages-6785–6797. doi:10.1074/jbc.RA120.013679.
- Beigel, J. H., Tomashek, K. M., Dodd, L. E., Mehta, A. K., Zingman, B. S., Kalil, A. C., Hohmann, E., Chu, H. Y., Luetkemeyer, A., Kline, S., Lopez de Castilla, D., Finberg, R. W., Dierberg, K., Tapson, V., Hsieh, L., Patterson, T. F., Paredes, R., Sweeney, D. A., Short, W., Touloumi, G., Lye, D., Ohmagari, N., Oh, M., Ruiz-Palacios, G., Benfield, T., Fätkenheuer, G., Kortepeter, M., Atmar, R., Creech, C., Lundgren, J., Babiker, A., Pett, S., Neaton, Burgess,T., Bonnett ,T., Green,M., Makowski, M., Osinusi, A., Nayak, S., Lane, H, N. Engl. J. Med, 2020, vol. 383(19), Pages-1813–1826. doi:10.1056/nejmoa2007764.
- Dangerfield, T. L., Huang, N. Z., Johnson, K. A, iScience, 2020, vol. 23(12), pp. 101849. doi: 10.1016/j.isci.2020.101849.
- Agostini, M. L., Andres, E. L., Sims, A. C., Graham, R. L., Sheahan, T. P., Lu, X., Smith, E. C., Case, J. B., Feng, J. Y., Jordan, R., Ray, A. S., Cihlar, T., Siegel, D., Mackman, R. L., Clarke, M. O., Baric, R. S., Denison, M. R. MBio, 2018, vol. 9(2), doi: 10.1128/mBio.00221-18.
- Jorgensen, S. C. J., Kebriaei, R., Dresser, L. D. Pharmacotherapy, 2022, vol. 40(7) , pp. 659–671. doi: 10.1002/phar.2429.
- Gordon, C. J., Tchesnokov, E. P., Feng, J. Y., Porter, D. P., Götte, M. J. Biol. Chem, 2020, vol. 295(15), pp. 4773–4779. doi:10.1074/jbc.AC120.013056.
- Shannon, A., Le, N. T. T., Selisko, B., Eydoux, C., Alvarez, K., Guillemot, J. C., Decroly, E., Peersen, O., Ferron, F., Canard, B. Antiviral Res, vol. 178, pp. 104793. doi:10.1016/j.antiviral.2020.104793.
- Yu, K. CrystEngComm, 2021, vol. 23(16), pp. 2923-2927. doi: 10.1039/d1ce00175b.
- Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., Shi, Z., Hu, Z., Zhong, W., Xiao, G. Cell Res, 2020, vol. 30(3), pp.269–271. doi: 10.1038/s41422-020-0282-0.
- Huang, L. F., Tong, W. Q. Adv. Drug Deliv. Rev,vol. 56(3), pp. 321–334. doi:10.1016/j.addr.2003.10.007.
- Siegel, D., Hui, H. C., Doerffler, E., Clarke, M. O., Chun, K., Zhang, L., Neville, S., Carra, E., Lew, W., Ross, B., Wang, Q., Wolfe, L., Jordan, R., Soloveva, V.,Knox, J., Perry, J., Perron, M., Stray, K. M., Barauskas, O., Feng, J., Xu, Y., Lee, G., Rheingold, A., Ray, A., Bannister, R., Strickley, R., Swaminathan, S., Lee, W., Bavari, S., Cihlar, T., Lo, M., Warren, T., Mackman, R. J. Med. Chem, 2017, vol. 60(5), pp. 1648–1661.
- doi: 10.1021/acs.jmedchem.6b01594.
- Parwani, A. (2022). Determinants of Working Capital Management: Evidence From Pharmaceutical Company of India –Cipla. October 2021.
- Multiple toxicities: 2 case reports. 2021. vol. 1, pp. 9954.
- Banik, N., & Chakraborty, D. Economic and Political Weekly, 2021 vol. 56(35), pp. 19–22.
- Organization, W.H, WHO Drug Infromation 2020, vol.34, pp. 862-876.
- Angel Pineiro, James Pipkin, Rebeca Gracia – Fandino, vol. 346, doi.org/10.1016/j.molliq.2021.117157.
- V.V.Tkach, M.Kushnir, S.C. de Oliveira, J.Ivanushko, A.V. Velyka, A.F. Molodianu, P.I. Yagodynets, Z.O. Karmosh, L.Yaz dos Reis, ,v, Luganska, vol.11, issue 2, 2021, 9201-9208, doi,org/10,3326/BRIAC112.92019208,
- Spinner, C. D., Gottlieb, R. L., Criner, G. J., Arribas López, J. R., Cattelan, A. M., Soriano Viladomiu, A., Ogbuagu, O., Malhotra, P., Mullane, K. M., Castagna, A., Chai, L. Y. A., Roestenberg, M., Tsang, O. T. Y., Bernasconi, E., Le Turnier,P., Chang, S. C., Sengupta, D., Hyland, R. H., Osinusi, A. O., Cao, H., Blair, C., Wang, H., Gaggar, A., Brainard, D., McPhail, M., Bhagani, S., Ahn, M., Sanyal, A., Huhn, G., Marty, F. M, JAMA -J. Am. Med. Assoc, 2020, vol. 324(11), pp.1048–1057. doi:10.1001/jama.2020.16349.
- Sun, D. AAPS J, 2020, vol. 22(4), pp.77. doi:10.1208/s12248-020-00459-8.
- Bhardwaj, Dr. Santosh. (2015). A Review: HPLC Method Development and Validation. International Journal of Analytical and Bioanalytical Chemistry, 2015; 5(4): 76-81.
- Organization, W.H WHO Drug Information 2020vol. 34, WHO Drug Information (2021), pp. 862-8764 [full issue].
- W.J. Umstead ,The chiral separation of remdesivir and several of its key starting materials ,LC-GC North America, 39 (6) (2021), pp. 291-293.
- B.A. Alden, P. Christensen, D. Foley, L.J. Calton, S. Barnes, G. Gallo, M.A. Lauber,Comprehending COVID-19: Mixed-Mode Chromatography for Ion Pairing Free LC-MS of Remdesivir and Remdesivir Triphosphate.Waters Corporation (2021).
- H.R. Reddy, S.R. Pratap, N. Chandrasekhar, S.Z.M. Shamshuddin. A novel liquid chromatographic method for the quantitative determination of degradation products in remdesivir injectable drug product J. Chromat. Sci. (2021)
- V. Patel, N. Tiwari, K. Patel, Stability indicating RP-HPLC method development and validation for the estimation of Remdesivir in API form. World J. Pharm. Pharmaceut. Sci., 10 (6) (2021), pp. 1544-1551
- Pasupuleti, R. R., Tsai, P. C., Ponnusamy, V. K., Pugazhendhi, A. Process Biochem, 2021, vol. 102, pp. 150–156. doi: 10.1016/j.procbio.2020.12.014.
- Ackermann, B., Berna, M., & Murphy, A. Curr. Top. Med. Chem, 2005, vol. 2(1), pp. 53–66. doi: 10.2174/1568026023394605.
- Tiller, P. R., Romanyshyn, L. A., Neue, U. D. Anal. Bioanal. Chem, 2003, vol. 377(5), pp. 788–802. doi: 10.1007/s00216-003-2146-0.
- Murphy, A. T., Berna, M. J., Holsapple, J. L., Ackermann, B. L. Rapid Commun. Mass Spectrom, 2005, vol. 16(6), pp. 537–543. doi:10.1002/rcm.606..
- Niessen, W. M. A. J. Chromatogr. A, 2003, vol. 1000(1-2), pp. 413–436. doi: 10.1016/S0021-9673(03)00506-5.
- Korfmacher, W. A. Drug Discov. Today, 2005, vol. 10(20), pp. 1357–1367. doi:10.1016/S1359-6446(05)03620-2.
- Xiao, D., John Ling, K. H., Tarnowski, T., Humeniuk, R., German, P., Mathias, A., Chu, J., Chen, Y. S., van Ingen, E. Anal. Biochem, 2021, vol. 617, pp. 114118. doi:10.1016/j.ab.2021.114118.
- Suresh Kumar, J. N., Keerthana, M., Varsha, N., Asha, P., Kumari, T. S., Babu, P. V., Prasad, B. S. Certif. J. 1559 World J. Pharm. Res. SJIF Impact Factor, 2022, vol. 11, pp. 1560. Doi. 10.20959/wjpr20224-23628.
- Habler, K., Brügel, M., Teupser, D., Liebchen, U., Scharf, C., Schönermarck, U., Vogeser, M., Paal, M. J. Pharm. Biomed. Anal, 2021, vol. 196. doi: doi.org/10.1016/j.jpba.2021.113935.
- Dadinaboyina, S. B., Yerra, N. V., Adimoolam, B. M., Parsa, S., Bathini, N. B., Thota, J. R. New J. Chem, 2021, vol. 45(16), pp. 7217–7224. doi: 10.1039/d1nj00160d.
- Skaggs, C., Zimmerman, H., Manicke, N., Kirkpatrick, L. J. Mass Spectrom. Adv. Clin. Lab, 2022, vol. 25,pp. 27–35. doi: 10.1016/j.jmsacl.2022.06.001.
- P.DU, G. Wang, S.Yang, P.Li, I. Liu Anal. Bioanal Chem., 413 (23) (2021) pp. 5811-5820.
- Organization, W.H, WHO Drug Information 2020, vol.34, pp. 862-876.
- V. Avantaneo, Ade Nicolo, J.Cusato, M. Antanucci, A. Manca, A. Palermiti, C. Waitt, S. Walimbwa, M. Lamorde, G. di Perri, A. D; Avolio, J. Antimicrochemother, 75 (7) (2020), pp.1772-1777, vol. 75, issue 7, doi,org/10.1093/jac/dkaa152.
- J.C.Alvarez, P. Moine, I.Etting, D.Annane, I.A.Larbi, Clinical Chemistry and Laboratory Medicine Journal, doi.org/10.1515/cclm-2020-0612.
- Wenjuan Hu, Lu Chang, Changqiang Ke, Yuanchao Xie, Jingshan Shen, Bo Tan, Jia Liu, vol.1994, doi.org/10.1016/j.pba.2020.113806
- Deqing Xiao, Kah. Hiing John Ling, Thomas Tarnowski, Rita Humeniuk, Polina German, Anita Mathias, Jasper Chu, Yuan-Shek Chan, Eric Van Ingen, vol.617, doi.org/10.1016/j.ab.2021.114118.
- Santodh Y. Gandhi, Barkha G.Kapoor, Journal of Drug Delivery and Therapeutics, dx.doi.org/10.2270/jddt.v914-5.3230.
- Ibrahim Bulduk, Erten Akbelvol. 15 issue, doi.org/10.1080/16583655.2021.1991737.
- Nguyen, R., Goodell, J. C., Shankarappa, P. S., Zimmerman, S., Yin, T., Peer, C. J., Figg, W. D. J. Chromatogr. B Anal. Technol. Biomed. Life Sci, 2021, vol. 1171,pp. 122641. doi. 10.1016/j.jchromb.2021.122641.
- Awady, M. El, Elmansi, H., Belal, F., Shabana, R. J. Fluoresc, 2022, vol. 32(5), pp. 1941-1948. doi: 10.1007/s10895-022-02998-z.
- Attia, T. Z., Boushra, J. M., Abdel Hakiem, A. F., Lashien, A. S., Noureldeen, D. A. M. Luminescence, 2022, vol. 37(7), pp. 1192–1199. doi: doi.org/10.1002/bio.4274.
- Pawar, N., Jalwal, P., Bahmani, K. J. Med. Pharm. Allied Sci, 2022, vol. 11(2), pp. 4603–4606. doi: 10.55522/jmpas.V11I2.2265.


 Tanvi Kamble*
Tanvi Kamble*
 Monali Khatake
Monali Khatake
 Nikita Pabale
Nikita Pabale
 Mansi Shelke
Mansi Shelke
 Vishweshwari Bhagat
Vishweshwari Bhagat

 10.5281/zenodo.14243862
10.5281/zenodo.14243862