Abstract
Alzheimer's disease (AD) and Parkinson's disease (PD) are among the most prevalent neurological disorders on a global scale, with significant impacts on cognitive functions, behavior, and memory. The genes apolipoprotein E (APOE), synuclein alpha (SNCA), and parkin RBR E3 ubiquitin-protein ligase (PRKN) are particularly important in the onset of these diseases. The activity and gene expression of these genes can be altered by genetic variations, based on prior research on them. To improve our understanding of potential damaging variants, our study used a variety of bioinformatics methods. Gene-to-gene or protein-protein interaction (PPI) study, docking analysis, and protein structure prediction were included as significant methods. Single-nucleotide variant statistics for the APOE, SNCA, and PRKN genes, which are linked with brain disorders, are offered in the GnomAD database, and UniProt evaluate sequences are accessible in FASTA format. These genes were studied using homology modeling by Swiss modeling and refinement was done by Galaxy Refine. We used the STITCH servers to study PPI, while the GeneMANIA tool was used to study gene-gene interactions. The potential inhibitors thought to possess a role in the onset of the disorder were found and evaluated using a molecular ligand docking method in the present study. Amphimedine, Deoxyamphimedine, and Quercetin were identified; all three have shown significant binding affinities, and these particular protein-ligand interactions offer significant insight for future studies. Despite these drawbacks, our analysis provides significant findings that will serve as an effective basis for future in vitro and in vivo research on heritable brain syndromes.
Keywords
APOE, SNCA, PRKN, STITCH, GENE MANNIA, SWISS model, GnomAD, PPI
Introduction
The most dominant forms of neurological conditions are Alzheimer's disease (AD) and Parkinson's disease (PD) in the world's population. Parkinson’s disease affects roughly 6 million individuals, while Alzheimer’s disease affects over 50 million people [1, 2]. In the year 2050, the projected population is expected to reach 152 million, with an annual growth rate of 5% [3, 4]. PD is the 2nd prevalent dementia after dementia, affecting 1-2% of persons over 65 and 4-5% of those over 85 [5]. Lewy bodies are a neuropathological hallmark of PD and are predominantly made up of the protein SNCA. The formation and deposit of beta-amyloid peptide into lesions in the fluid brain tissue, followed by hyperphosphorylated or tau in intracellular neurofibrillary tangles [6], named after the German psychiatrist Alois Alzheimer, AD is observed [7]. Although PD is referred to as another age-related neurotoxic disease, it is recognized clinically by rigidity, tremor, and loss of activity, as well as neuropathologically by gradual neuronal loss of the spinal nerve [8]. Mutations in the PRKN gene represent the primary cause of autosomal-recessive juvenile PD [9]. The ABCA1, APOE, MAPT, and SNCA genes are typically related to AD and are related to cognitive function in PD pathogenesis, and SNCA mutations are identified in familial forms of the disease [10]. Globally, dementia is expected to cost $1 trillion annually, placing a significant strain on people, families, and communities [11, 12]. While there is no FDA-approved treatment for AD, there are drugs that can help with the symptoms [13]. A genetic mutation in the LRRK2, PARK2, PARK7, PINK1, or SNCA gene can cause family-linked PD, which affects around 15% of people. The underlying causes of PD involve the presence of intracellular Lewy body inclusions within neurons and the loss of dopaminergic neurons in the substantia nigra pars compacta (SNpc) [10]. Protein aggregation and gene mutation are major causes of dopamine neuron degeneration. However, the exact cause of dopamine degradation is unknown, and several atypical PD family types are linked to specific genetic loci, knowing the disease's biology [14]. Certain genes, such as the Apolipoprotein E (APOE) gene, affect later-onset dementia, which is more common. An increased risk for this disorder varies with APOE gene variation, notably APOE4 [15]. Familial PD is related to specific mutations in genes such as Synuclein Alpha (SNCA) [16, 17] and Parkin protein (PRKN) [18]. The APOE gene encodes a 317-aminoacid protein that is found on chromosome 19q13.32 with total exons 6. The three common variations, ?2, ?3, and ?4, each produce a single amino acid change in the APOE protein. Specifically, APOE ?4 is linked with a higher risk of dementia, whereas APOE ?2 with lower risk factors [19]. The variance in the APOE genotype affects beta-amyloid buildup in the brain's cortex and its blood vessels appears to be the basis of the APOE genotype's impact on the emergence of various disorders [20]. Significant evidence ties the APOE gene to the risk of dementia and has been studied in various neurological disorders [21, 22]. The SNCA gene contains alpha-synuclein and is found on chromosome 4q22.1, with 12 exons [16]. The initial association between ?-synuclein and PD was established in 1997 when a coding SNP A to T at position 53 in SNCA was linked to autosoma dominant Parkinsonism [23]. Various researchers found that large SNCA aggregates led to the etiology of an array of neurological conditions, including PD, dementia (AD), multiple systems atrophy (MSA), and dementia with Lewy bodies (DLB) [24]. The protein alpha-synuclein, which has 140 amino acids, is a member of the same protein family as the ?- and ?-synucleins [25]. The first mutation in the SNCA gene, which codes for the ASN protein, and the first mutation found in PD, was reported in 1990 [26]. The first mutation in the Parkin protein (PRKN) gene discovered in 1998 [10, 27]. Biallelic PKRN gene variants are known to be responsible for autosomal recessive Parkinson's disease (PD). However, the role of monoallelic PRKN alterations as a risk factor for PD is still uncertain [28]. Parkin, a 465 amino acid autoinhibited E3 ubiquitin ligase, is encoded by PRKN comprising13 exons and is mapped on human chromosome 6q26 [29, 30]. A global study is being carried out on the genetics of the neurological condition, and reports of variants are emerging. The two most often found genes, PINK1 and PARK2, are both related to autosomal recessive forms of PD [31]. Moreover, In silico studies have a substantial impact on the recognition of potential SNPs because they are quick, economical, and can aid with future genetic research [32]. The objective of this study is to forecast the 3D structures of the APOE, SNCA, and PRKN proteins using the SWISS model, which relies on PyMOL serves to display molecular crystal structures. In this study, computational methods including GeneMANIA and STITCH analysis were employed to find the gene-gene and PPI interactions. Furthermore, protein-ligand docking was used to explore the associations of APOE, SNCA, and PRKN with ligands. Despite its drawbacks, our study generated insights that will be useful for future lab-based and animal studies regarding gene-associated neurological disorders.
MATERIALS AND METHOD
The comprehensive methodology of this current
paper is illustrated in Figure 1.
Analysis of variants from the GnomAD database
The protein SNP data for the APOE, SNCA, and PRKN genes acquired from GnomAD are available at https://gnomad.broadinstitute.org. A worldwide collaboration of researchers established the Genome Aggregation Database Consortium, commonly referred to as GnomAD. Their goal was to collect and standardize exome and genome sequencing data obtained from various extensive sequencing initiatives and furnish data summaries to the scientific community [33, 34]. Previously, this initiative was recognized as the Exome Aggregation Consortium (ExAC). The reference genome employed for sequence alignment was GRCh37/hg19, and the alignment process was executed using the GATK tool [35].

Figure 1. Methodology of the present paper.
Data Mining and sequence reterival
The APOE, SNCA, and PRKN sequences were obtained in the FASTA format from the Universal Protein Resource (UniProt) at http://www.uniprot.org. The APOE from Homo sapiens (P02649), SNCA (P37840), and PRKN (O60260) are about of these proteins [36]. The APOE, SNCA, and PRKN gene data information was retrieved from the NCBI database at https://www.ncbi.nlm.nih.gov [5]. Utilizing computational modeling methods, the sequences of the aforementioned proteins were analyzed, and 3D models were built. The protein data taken into account for this investigation is displayed in Table 1.
Biophysical and Visualization Analysis
GeneMANIA available at https://genemania.org is a web server that develops hypotheses on gene function, aspects into gene lists, and ranks genes for functional testing [37]. The website GeneMANIA may identify new genes linked to a set of input genes by utilizing a vast repository of biological correlation information. Protein interactions with genes, coexpression, methods, and colocalization are all examples of interaction statistics [38, 39]. To study how proteins interact, the STITCH server, sited at http://stitch.embl.de was used. For quick access to verified theoretical and experimental interactions of the required protein, this database offers an essential evaluation and integration of protein-protein interactions [40, 41].
Prediction of protein models
The protein sequences that were obtained were used as query sequences in comparative modeling. The 3D structures of APOE, SNCA, and PRKN were predicted using SWISS-MODEL are given at http://swissmodel.expasy.org, which also included integrated external data sources like UniProt, STRING analysis and used to predict the protein crystal templates [42]. The refinement is done by Galaxy Refine available at https://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE. UCSF Chimera is installed from http://www.cgl.ucsf.edu/chimera [43], Discovery Studio was downloaded from https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio [44], PyMOL available at https://pymol.org [45] is a visualization tool of 3D structure.
Model evaluation by SAVES server
The core stability and reliability of the modeled structures of the APOE, SNCA, and PRKN were assessed using several ways. By counting the residues in the permitted zones of the Ramachandran plot, the stereochemical quality of the model was evaluated using SAVES algorithms available at https://saves.mbi.ucla.edu [46]. The derived protein structure was reevaluated using QMEAN Z-scores from the QMEAN tools available at http://swissmodel.expasy.org/docs/structure_assessment for its dependability and model quality. When evaluating a protein's 3D structure, SAVES employs six modules, including PROCHECK, 3D Verify, and ERRAT, to ensure the quality of the entire 3D model [47, 48]. A Ramachandran plot of psi-phi torsion angles are produced by the PROCHECK, with over 90% of residue in the most favored regions likely to be in acceptable quality. The Ramachandran plot, which displays how residues behave in zones where they are allowed and not allowed [49]. PROCHECK analyzes both the total structural geometry and the geometry of each distinct residue to ascertain the stereochemical quality of a protein structure [50]. ERRAT evaluates the overall model quality based on the probability distribution of non-bonded interactions among a variety of atoms relying on shared atomic interactions [48, 51, 52].
Virtual screening of protein-ligand docking
The SDF files for the identified ligands, including Amphimedine, Benazepril, Captopril, Curcumin, Deoxyamphimedine, Emetine, Fibrinogen, Hyaluronic Acid, Lacosamide, and Quercetin, were made available by the Protein Data Bank (PDB), PubChem, and EMBL-EBI databases. More details regarding these substances and their biological features may be found in publications. PYMOL was used to convert the SDF file containing the ligand structures to the PDB format [45]. The variety of potential targets for drug discovery has increased for the study of the human genome. High-throughput crystallography, protein purification, and NMR spectroscopy (nuclear magnetic resonance) were all used to offer structural data on protein complexes [53, 54]. Such advances produced molecular docking also referred to as computer-assisted drug design (CAD). Molecular docking employs both ligand-based and structure-based approaches [55]. In this study, molecular docking of known organic antagonists was done to identify the most effective ligand, which may one day serve as the basis for developing drugs against psychiatric diseases caused by APOE, SNCA, and PRKN. Several algorithmic methods were developed and are frequently used for protein-ligand docking, especially if looking for drugs based on small, well-structured molecules. The objectives of molecular docking were to identify ligand-protein interactions and possible ligands [56]. Using the PyRx tool available at https://pyrx.sourceforge.io we docked all of the chosen ligands with targeted proteins. For virtual ligand screening, the Lamarckian genetic algorithm (LGA), which integrates AutoDock and AutoDock Vina, was used [57-59]. Each ligand's top ten exclusive values were calculated, with the active parameters set to the center's (XYZ axis) grid size. The PDB files were transformed into PDBQT format using AutoDock tools, following which the binding affinities were computed [60]. For virtual screening, the 2D and 3D interactions of ligands with proteins were visualized using Discovery Studio. They displayed a docked ligand's bonding lengths, hydrogen bond interactions, hydrophobic interactions, and the size and position of its bonding sites [61, 62].

Figure 2. Mutations in the APOE, SNCA, and PRKN genes in Homo sapiens were identified.
RESULTS AND DISCUSSION
SNP analysis of the target gene
This study used the gnomAD database to observe polymorphic variants of the APOE, SNCA, and PRKN genes and examine potential risk factors. The findings showed typical mutations, 188 of which are missense, 87 synonymous regions, and 6 in pLoF were detected in the APOE gene. The data revealed that 61 are missense mutations, 31 are in the synonymous region in the SNCA gene and 310 are missense, 124 are synonymous and 14 pLoF in the PRKN gene. However, current research indicates such variations are most likely linked to neurological and mental disorders. To ascertain the importance of these changes in disease progression, analyses of function are thus required. Figure 2 shows that mutations in the APOE, SNCA, and PRKN genes of Homo sapiens have been found.
Retrieval of FASTA sequence
The selection of the APOE, SNCA, and PRKN genes for this study was based on their involvement in the initial stages of neurodegenerative diseases, specifically Alzheimer's disease and Parkinson's disease. Sequences in FASTA format were obtained from UniProt and subsequently employed for in-depth analysis. The protein sequences under evaluation are outlined in Table 1.
Gene-gene interaction
The APOE gene relates to 20 different genes, namely APOC1, LIPC, APOA1, APOA2, APOA4, APOA5, VLDLR, LDLR, APOC2, APOC3, APOB, LRP1, LRP8, SORL1, APP, LDLRAP1, CNTF, ZIC1, MAPT, and TMCC2. APOE co-expresses 17 genes, including APOC1, APOA1, APOC3, APOC2, APOA2, APOB, LIPC, APOA4, APP, APOA5, CNTF, MAPT, SORL1, LRP1, LDLR, and LRP8. APOE also shares a domain with eight other genes; VLDLR, LDLR, LRP8, LRP1, SORL1, APOA5, APOA1, and APOA4. These data indicate that APOE and the associated genes may be functionally related and may share 14 similar biological pathways. These findings provide a foundation for further research into the biological activities of APOE and its possible involvement in related pathways, as seen in Figure 3 by showing that the SNCA gene interacts with several Synuclein families, including CHMP5, SNCAIP, MARK1, AK1, KLK6, TUBB3, SNCG, CNP, MYO5A, TPPP, PPP2CA, STX1A, SNCB, HSPA9, MT-ND5, PARK7, APCS, UCHL-1, SLC6A3, and SRCIN1. Figure 4 depicts the 14 coexpression genes and their contributions to achieving comparable activities or sharing four similar protein domains.

Figure 4. Gene-gene interaction network of SNCA protein illustrated by GeneMANIA.

Figure 5. Gene-gene interaction network of PRKN protein illustrated by GeneMANIA.
Protein-Protein Interaction
String discovered that APOE, SNCA, and PRKN proteins interact with other proteins involved in neuroglial endeavors, and Figure 6 depicts other interactions with proteins. To anticipate interactions and elucidate their molecular consequences, the STITCH and STRING databases were employed to build a protein-protein interaction network (PPIN) and to incorporate chemical compounds into the human protein network. In this network, edges represent different types of interactions, chemicals are depicted as tablets, proteins are represented as circles, and the numerical value "degree" indicates the level of activity. Larger nodes correspond to higher degree values, and as the degree value increases, the color shifts from light green to yellow. The network, comprising nodes and edges, is presented in Table 2. The network nodes act as proxies for the genes that code for the protein targets. The connections between the different nodes show how the relevant proteins interact with the bioactive substances.


Figure 6. The STITCH 5.0 displayed a chain of associations between bioactive molecules and the protein targets a) APOE, b) SNCA, and c) PRKN. The circles stand in for proteins, whereas pill-shaped nodes represent compounds.
Table 2. Protein-protein network stats by STITCH analysis

???????
Homology protein modeling by SWISS Model
For comparative homology modeling the generated sequences were selected of at least >30% similarities and identities. The protein modeling is achieved by the SWISS model of target protein APOE, SNCA, and PRKN. The amino acid of APOE was 317, we got 15 templates with 93.06% STML ID P0DKW7.1. A (Range:1-317; coverage: 1.00), Alphafold DB model of APOE_PLEMO (Organism: Dusky titi monkey) identification for the query sequence. For the 140 amino acid of SNCA, we got 10 templates with 99.29% STML ID P61139.1.A (Range:1-140; coverage: 1.00), Alphafold DB model of SYUA_ERYPA (Organism: Red guenon) identification for the query sequence. For the 465 amino acids of PRKN, we got 50 templates with 99.01% STML ID 5c1z.1.A (Range:1-465; coverage: 0.87), model of Parkin (Organism: UbIRORBR) identification for the query sequence. To learn how mutations drastically alter the stability of proteins, we modeled the three-dimensional structure of the APOE, SNCA and PRKN protein using the template PDB ID. The results of using a template with quality and PyMoL to create a model are displayed in Figure 7. UCSF Chimera [43], Discovery Studio [44], PyMOL [45] is a visualization tool of 3D structure. Validation of the modeled framework was performed by SAVES and RAMACHANDRAN plot evaluation was used to examine the secondary structure. All constraints imposed by potential energy calculations were respected by the resulting structure. In the RAMACHANDRAN plot, a significant portion of the amino acid residues within the target proteins were located within a highly favorable region, as visualized in Figure 7. You can access the detailed predicted results in Table 3.
Figure 7. Predicted models of APOE, SNCA and PRKN gene, validated by QMEAN and Ramachandran plot.

Molecular Docking by PyRx
To discover ligand-protein interactions, molecular docking was used by PyRx tool, we docked all of the selected ligands with APOE, SNCA, and PRKN proteins. They created ten distinct conformations for each ligand, which are characterized by binding affinity (-Kcal/mol).
The docking results of ligands indicate that these binding affinities relate to their level of activity, and all 10 small molecules with binding affinities are given in Table 4 and 3D and 2D representations of docking scores are visualized by Discovery Studio as shown in Figure 8.
Table 3. Statistics of Ramachandran plot and analysis of the models from Swiss model

We selected 3 compounds with strong binding affinities, notably Amphimedine, Deoxyamphimedine and Quercetin, and docked them with all 3 of our native protein complexes such as APOE, SNCA and PRKN further to investigate their interactions as depicted in Table 5.
Table 4.
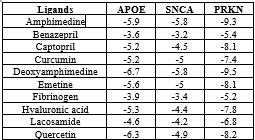

Figure 8. 2D and 3D representation of all docking interactions
A Deoxyamphimedine and quercetin ligand was fixed in the APOE binding pocket sites by forming the conventional hydrogen bond with residues GLU 106, TYR92, GLN181; and hydrophobic interactions (Alkyl, pi-alkyl, cation, anion, halogens) with ALA 109, LYS113, ARG110, GLU114, LEU177, ALA178, GLN174, TRP38, THR36, GLU37, SER40, and GLN42. The SNCA docked with Amphimedine and Deoxyamphimedine by showing hydrogen bond with residues GLU20; as hydrophobic interactions with GLU13, ALA17, VAL16, LYS12, ILE88, ALA85, THR92, VAL95, LYS96, PHE94, and ALA89. Moreover, PRKN protein shows PHE264, HIS265, LEU272, GLU321, and GLY338 are vander wall forces; and LEU307, ARG271, LEU287, PHE277, THR270, TYR267, VAL269, LEU266, ALA339, LEU358, GLN317, LEU331, SER10, SER9, ASN8, and ARG314 residues showing hydrophobic interactions as depicted in Table 5. The interacting residues obtained from docking are presented in Figure 8. According to the score, polymorphisms not only alter the protein's conformation because of modifications to the number of interacting residues but also because of alterations in hydrogen bonds and hydrophilic associations.
Table 5. Interacting residues of APOE, SNCA and PRKN with ligands including their binding residues and hydrophobic interactions.

CONCLUSION
The genes responsible for encoding APOE, SNCA, and PRKN plays pivotal roles in the development of Alzheimer's and Parkinson's diseases. To gain deeper insights into the potential pathological aspects associated with these genes, we employed a comprehensive array of bioinformatics methodologies to construct a structural model. Our approach encompassed gene-to-gene and protein-protein interaction (PPI) analysis, docking analysis, and protein structure prediction. For gene modeling, we utilized homology modeling via the Swiss modeling platform, followed by refinement using Galaxy Refine. In examining PPI, the STITCH servers were employed, while the GeneMANIA tool facilitated the investigation of gene-gene interactions. Furthermore, we evaluated potential disease-associated inhibitors through a molecular ligand docking technique. Notably, we identified three compounds, namely Amphimedine, Deoxyamphimedine, and Quercetin, all of which exhibited substantial binding affinities. These specific protein-ligand interactions provide valuable insights for future research endeavors. Furthermore, these identified therapeutic compounds may prove to be potent and effective molecules capable of interacting with neuronal proteins, making them promising candidates for drug development.
ACKNOWLEDGMENT
No acknowledgment.
AUTHOR CONTRIBUTION
The work has been reviewed by all authors, who have given their approval for publishing in the current form.
CONFLICT OF INTEREST
Authors don't have any competing interests.
REFERENCES
- Dutta, S., et al., Biomarkers for parkinsonian disorders in CNS-originating EVs: promise and challenges. Acta Neuropathologica, 2023. 145(5): p. 515-540.
- Skidmore, S. and R.A. Barker, Challenges in the clinical advancement of cell therapies for Parkinson’s disease. Nature Biomedical Engineering, 2023: p. 1-17.
- Caligiore, D., F. Giocondo, and M. Silvetti, The Neurodegenerative Elderly Syndrome (NES) hypothesis: Alzheimer and Parkinson are two faces of the same disease. IBRO Neuroscience Reports, 2022.
- Dorsey, E., et al., The emerging evidence of the Parkinson pandemic. Journal of Parkinson's disease, 2018. 8(s1): p. S3-S8.
- Ni, X., et al., Structural insights into ?-synuclein fibril polymorphism: effects of Parkinson's disease-related C-terminal truncations. Journal of molecular biology, 2019. 431(19): p. 3913-3919.
- Querfurth, H.W. and F.M. LaFerla, Alzheimer's disease. New England Journal of Medicine, 2010. 362(4): p. 329-344.
- Klostermeier, S., et al., Exploring the Skin Brain Link: Biomarkers in the Skin with Implications for Aging Research and Alzheimer’s Disease Diagnostics. International Journal of Molecular Sciences, 2023. 24(17): p. 13309.
- Abaza, A., et al., Parkinson’s Neuropathology Puzzle: A Systematic Review Uncovering the Pathological Culprits Behind the Neurological Disease. Cureus, 2023. 15(8).
- Li, H.-x., et al., A homozygous PRKN-associated juvenile Parkinson's disease with pregnancy in China. Frontiers in Neurology, 2023. 14: p. 1103164.
- Oczkowska, A., et al., Mutations in PRKN and SNCA genes important for the progress of Parkinson’s disease. Current genomics, 2013. 14(8): p. 502-517.
- Yiannopoulou, K.G. and S.G. Papageorgiou, Current and future treatments in Alzheimer disease: an update. Journal of central nervous system disease, 2020. 12: p. 1179573520907397.
- Livingston, G., et al., Dementia prevention, intervention, and care: 2020 report of the Lancet Commission. The Lancet, 2020. 396(10248): p. 413-446.
- Breijyeh, Z. and R. Karaman, Comprehensive review on Alzheimer’s disease: Causes and treatment. Molecules, 2020. 25(24): p. 5789.
- Selvaraj, S. and S. Piramanayagam, Impact of gene mutation in the development of Parkinson's disease. Genes & diseases, 2019. 6(2): p. 120-128.
- Rohn, T.T., et al., The potential of CRISPR/Cas9 gene editing as a treatment strategy for Alzheimer’s disease. Journal of Alzheimer's disease & Parkinsonism, 2018. 8(3).
- Siddiqui, I.J., N. Pervaiz, and A.A. Abbasi, The Parkinson Disease gene SNCA: Evolutionary and structural insights with pathological implication. Scientific reports, 2016. 6(1): p. 24475.
- Hashimoto, M. and E. Masliah, Alpha?synuclein in Lewy body disease and Alzheimer's disease. Brain pathology, 1999. 9(4): p. 707-720.
- Elsayed, I., et al., Mapping the Diverse and Inclusive Future of Parkinson’s Disease Genetics and Its Widespread Impact. Genes, 2021. 12(11): p. 1681.
- Stefanidou, M., et al., Bi-directional association between epilepsy and dementia: The Framingham Heart Study. Neurology, 2020. 95(24): p. e3241-e3247.
- Yang, L.G., et al., Apolipoprotein E in lipid metabolism and neurodegenerative disease. Trends in Endocrinology & Metabolism, 2023.
- Verghese, P.B., J.M. Castellano, and D.M. Holtzman, Apolipoprotein E in Alzheimer's disease and other neurological disorders. The Lancet Neurology, 2011. 10(3): p. 241-252.
- Pocevi?i?t?, D., et al., Increased plasma and brain immunoglobulin A in Alzheimer’s disease is lost in apolipoprotein E ?4 carriers. Alzheimer's Research & Therapy, 2022. 14(1): p. 117.
- Polymeropoulos, M.H., et al., Mutation in the ?-synuclein gene identified in families with Parkinson's disease. science, 1997. 276(5321): p. 2045-2047.
- Wang, Q., et al., SNCA gene polymorphism may contribute to an increased risk of Alzheimer's disease. Journal of clinical laboratory analysis, 2016. 30(6): p. 1092-1099.
- Qi, Z., et al., Influence of Cholesterol on the Membrane Binding and Conformation of ?-Synuclein. The Journal of Physical Chemistry B, 2023. 127(9): p. 1956-1964.
- Priya, M., et al., Quantum Chemical Investigation, Drug-Likeness and Molecular Docking Studies on Galangin as Alpha-Synuclein Regulator for the Treatment of Parkinson’s Disease. Chemistry Africa, 2023. 6(1): p. 287-309.
- Yoshino, H., et al., Genotype-phenotype correlation of Parkinson's disease with PRKN variants. Neurobiology of Aging, 2022. 114: p. 117-128.
- Lubbe, S.J., et al., Assessing the relationship between monoallelic PRKN mutations and Parkinson’s risk. Human molecular genetics, 2021. 30(1): p. 78-86.
- Seike, N., et al., Genetic variations and neuropathologic features of patients with PRKN mutations. Movement Disorders, 2021. 36(7): p. 1634-1643.
- Castelo Rueda, M.P., et al., Frequency of Heterozygous Parkin (PRKN) Variants and Penetrance of Parkinson's Disease Risk Markers in the Population-Based CHRIS Cohort. Frontiers in neurology, 2021. 12: p. 706145.
- Goldstein, O., et al., Novel variants in genes related to vesicle-mediated-transport modify Parkinson's disease risk. Molecular Genetics and Metabolism, 2023. 139(2): p. 107608.
- Bakhit, Y.H.Y., et al., In silico analysis of SNPs in PARK2 and PINK1 genes that potentially cause autosomal recessive Parkinson disease. Advances in Bioinformatics, 2016. 2016.
- Karczewski, K.J., et al., The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 2020. 581(7809): p. 434-443.
- Koch, L., Exploring human genomic diversity with gnomAD. Nature Reviews Genetics, 2020. 21(8): p. 448-448.
- Souza Junior, M.L.F., J.V. de Sousa, and J.F. Guerreiro, Analysis of coding variants in the human FTO gene from the gnomAD database. Plos one, 2022. 17(1): p. e0248610.
- Nörenberg, D., et al., Molecular analysis of a novel Toll/interleukin-1 receptor (TIR)-domain containing virulence protein of Y. pseudotuberculosis among Far East scarlet-like fever serotype I strains. International Journal of Medical Microbiology, 2013. 303(8): p. 583-594.
- Mustafa, M.I., et al., Extensive in silico analysis of ATL1 gene: discovered five mutations that may cause hereditary spastic paraplegia type 3A. Scientifica, 2020. 2020.
- Chetta, M., et al., Rising roles of small noncoding RNAs in cotranscriptional regulation: in silico study of miRNA and piRNA regulatory network in humans. Genes, 2020. 11(5): p. 482.
- Barali?, K., et al., Potential genomic biomarkers of obesity and its comorbidities for phthalates and bisphenol A mixture: In silico toxicogenomic approach. Biocell, 2022. 46(2): p. 519-533.
- Ezaouine, A., et al., In Silico Prediction of the Bioactive Profile and Metabolites of Satureja nepeta in Diseases Related to the Excessive Production of Interleukin-6. Bioinformatics and Biology Insights, 2022. 16: p. 11779322221115665.
- Ahuja, K., et al., Comparative Analysis of Computational Methods used in Protein-Protein Interaction (PPI) Studies, in Computational and Analytic Methods in Biological Sciences: Bioinformatics with Machine Learning and Mathematical Modelling. 2023, River Publishers. p. 63-100.
- HIdayat, H., W. Haryadi, and T.J. RAHARJO, Three-dimensional structure modeling of a protease from lactic acid bacteria Leuconostoc mesenteroides K7 using automated protein homology analysis. Biodiversitas Journal of Biological Diversity, 2020. 21(7).
- Pettersen, E.F., et al., UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry, 2004. 25(13): p. 1605-1612.
- Adeniji, S.E., S. Uba, and A. Uzairu, In silico study for evaluating the binding mode and interaction of 1, 2, 4-triazole and its derivatives as potent inhibitors against Lipoate protein B (LipB). Journal of King Saud University-Science, 2020. 32(1): p. 475-485.
- Schrödinger, L., The PyMOL molecular graphics system, version 1.8. 2015, November.
- Wlodawer, A., Stereochemistry and validation of macromolecular structures. Protein Crystallography: Methods and Protocols, 2017: p. 595-610.
- Mahmud, Z., et al., Computational analysis of damaging single-nucleotide polymorphisms and their structural and functional impact on the insulin receptor. BioMed research international, 2016. 2016.
- Colovos, C. and T.O. Yeates, Verification of protein structures: patterns of nonbonded atomic interactions. Protein science, 1993. 2(9): p. 1511-1519.
- Hasnain, M.J.U., et al., Computational analysis of functional single nucleotide polymorphisms associated with SLC26A4 gene. PLoS One, 2020. 15(1): p. e0225368.
- Laskowski, R.A., et al., PROCHECK: a program to check the stereochemical quality of protein structures. Journal of applied crystallography, 1993. 26(2): p. 283-291.
- Rozario, L.T., T. Sharker, and T.A. Nila, In silico analysis of deleterious SNPs of human MTUS1 gene and their impacts on subsequent protein structure and function. Plos One, 2021. 16(6): p. e0252932.
- Santhoshkumar, R. and A. Yusuf, In silico structural modeling and analysis of physicochemical properties of curcumin synthase (CURS1, CURS2, and CURS3) proteins of Curcuma longa. Journal of Genetic Engineering and Biotechnology, 2020. 18(1): p. 1-9.
- Bajorath, J., Integration of virtual and high-throughput screening. Nature Reviews Drug Discovery, 2002. 1(11): p. 882-894.
- Jorgensen, W.L., The many roles of computation in drug discovery. Science, 2004. 303(5665): p. 1813-1818.
- Kuntz, I.D., et al., A geometric approach to macromolecule-ligand interactions. Journal of molecular biology, 1982. 161(2): p. 269-288.
- Halder, S.K., et al., A comprehensive study to unleash the putative inhibitors of serotype2 of dengue virus: insights from an in silico structure-based drug discovery. Molecular Biotechnology, 2022: p. 1-14.
- Dallakyan, S. and A.J. Olson, Small-molecule library screening by docking with PyRx, in Chemical biology. 2015, Springer. p. 243-250.
- Morris, G.M., R. Huey, and A.J. Olson, Using autodock for ligand?receptor docking. Current protocols in bioinformatics, 2008. 24(1): p. 8.14. 1-8.14. 40.
- Trott, O. and A.J. Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry, 2010. 31(2): p. 455-461.
- Ferrari, I.V. and P. Patrizio, Development and Validation Molecular Docking Analysis of Human serum albumin (HSA). bioRxiv, 2021.
- Afriza, D., W. Suriyah, and S. Ichwan. In silico analysis of molecular interactions between the anti-apoptotic protein survivin and dentatin, nordentatin, and quercetin. in Journal of Physics: Conference Series. 2018. IOP Publishing.
- Anwar, T., et al., Insilico modeling and analysis of small molecules binding to the PHLPP1 protein by molecular dynamics simulation. Pakistan Journal of Pharmaceutical Sciences, 2020. 33.


 Kainat Ramzan* 2
Kainat Ramzan* 2
 Ibtsam Bilal 2
Ibtsam Bilal 2











 10.5281/zenodo.13904186
10.5281/zenodo.13904186