Abstract
Microbial infections and rising antimicrobial resistance highlight the need for new treatments. Benzoxazine derivatives are promising candidates due to their diverse biological activities. This study synthesized and evaluated the antimicrobial properties of 2H-benzo[b][1,4] oxazin-3(4H)-one derivatives. The synthesis involved reacting 2-aminophenol with chloroacetic acid, followed by sulfonation with chlorosulfonic acid and subsequent nucleophilic substitution with aryl amines. The compounds were tested against E. coli, S. aureus, and B. subtilis, with compound 4e showing the highest activity across all strains. Molecular docking studies targeting E. coli DNA gyrase revealed that compound 4d had the strongest binding affinity, followed by compounds 4a, 4e, and 4f. Binding interactions with key amino acid residues were analyzed, providing insights for further optimization. Compound 4e’s broad-spectrum activity and high potency suggest its potential as a new antimicrobial agent, warranting further in vivo testing and toxicity evaluation.
Keywords
Benzoxazines, Benzoxazolinones, Molecular Docking etc.
Introduction
The increasing prevalence of microbial infections, exacerbated by the rise of antimicrobial resistance, poses a significant challenge to global public health. Traditional antibiotics are losing their effectiveness as bacteria and fungi evolve mechanisms to evade these treatments, necessitating the development of novel therapeutic strategies. Among the promising candidates are benzoxazine derivatives, a class of compounds that have garnered attention for their diverse biological activities and relative chemical simplicity.1-5 Benzoxazine belongs to a bicyclic system having an oxazine ring annulated to a benzene ring. Its two common derivatives, with and without a keto group, are known as 2H-1,4-benzoxazin-3-(4H)-one, and 3,4-dihydro-2H-1,4-benzoxazine, respectively shown fig no.1.

Figure 1: Benzoxazine belongs to a bicyclic system having an oxazine ring annulated to a benzene ring.
Benzoxazines, characterized by their fused oxazine ring structure, exhibit a range of pharmacological effects, including anti-inflammatory, anti-ulcer, antibacterial, and antifungal properties. These compounds primarily exist in the form of aglycones in living systems, where they function as defense chemicals against various organisms such as fungi, bacteria, insects, and weeds. This defensive role is underscored by their presence as natural plant toxins in economically important species within the Poaceae family (e.g., maize, wheat, and rye) and some dicotyledonous species from the Acanthaceae, Lamiaceae, Scrophulariaceae, and Ranunculaceae families. The biological relevance of benzoxazines extends to their analogues, which include cyclic hydroxamic acids, their corresponding lactams, and ring-contracted benzoxazolinones. These naturally occurring compounds are produced as secondary metabolites in plants and have been recognized for their potential antimicrobial properties. The structural simplicity and versatility of benzoxazine derivatives make them attractive candidates for synthetic modification, aimed at enhancing their efficacy as antimicrobial agents. This study focuses on the synthesis and evaluation of 2H-benzo[b] [1,4] oxazin-3(4H)-one derivatives for their antimicrobial activity. By exploring the chemical modifications of these compounds and assessing their effectiveness against various bacterial strains, we aim to identify potent new antimicrobial agents. The findings could contribute significantly to the development of novel treatments to combat resistant microbial infections, addressing a critical need in contemporary medicine.6-10
Experimental:
Chemicals and reagents: Chloroacetyl chloride,chloroform,2-aminophenol,TEBA, sodium, Bicarbonate,Synthesis pathway of Benzoxazine derivatives 4(a-g)wereobtained from DCS’s ARA College of pharmacy, Nagaon, Dhule(Loba chem-Mumbai, India). The Synthesis compounds were tested to confirm their identity and this included recording of FT-IR spectra. FT-IR spectra were recorded at ARACOP, Dhule. The sample was prepared as a KBrpellet for recording the spectra.
Step I:
Chloroacetyl chloride (72.2 mmol) in chloroform (5 mL) was added over 20 min to a suspension of 2-aminophenol (50.0 mmol), TEBA (50.0 mmol), and sodium bicarbonate (200 mmol) in chloroform (30 mL) at 0°C. The mixture was stirred for 1 hour, heated to 55°C for 16 hours, then concentrated and diluted with water. The precipitate was filtered, washed with water, dried under vacuum, and recrystallized from ethanol to yield 2H-benzo[b][1,4]oxazine-3(4H)-one.
Step II:
2H-benzo[b] [1, 4] oxazine-3(4H)-one (13.4 mmol) was added over 20 min to sulfurochloridic acid (10 mL) at 0°C and stirred for 1 hour. The mixture was poured onto ice, extracted with dichloromethane, dried over sodium sulfate, and concentrated to yield 3-oxo-3,4-dihydro-2H-benzo[b][1,4]oxazine-6-sulfonyl chloride.
Step III:
A solution of 3-oxo-3, 4-dihydro-2H-benzo[b] [1, 4] oxazine-6-sulfonyl chloride and different arylamines (0.463 mmol) in pyridine at 0°C was stirred at room temperature for 2 hours. The mixture was diluted with water, extracted with dichloromethane, washed with 6N HCl, dried over sodium sulfate, and concentrated under reduced pressure to obtain product 4a shown in fig no.2, and table no.1.11-20
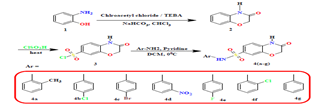
Figure 2: Synthesis pathway of Benzoxazine derivatives 4(a-g)
Table 1: Physical data for synthesized derivatives 4a to 4g:

Pharmacological Screening Procedure:
Antibacterial and Antifungal Activity (Disc Diffusion Assay):
Microorganisms:
Bacteria: Staphylococcus aureus (NCIM 2079), Escherichia coli (NCIM 2109), and Bacillus subtilis (NCIM 5433). Fungi: Aspergillus Niger (NCIM 545)
Preparation:
Grow pure cultures in nutrient broth (bacteria) and MGYP medium (fungi) at 37°C until turbidity matches 0.5 McFarland standards (2 x 10^8 CFU/ml). Adjust turbidity with sterile saline or broth.
Inoculation:
Dip a sterile cotton swab into the adjusted suspension, streak over nutrient agar plates evenly by rotating the plate approximately 60° each time.
Disc Preparation:
Prepare stock solutions (1000 ?g/ml) of compounds (4a–4g) in DMSO. Use 6 mm sterile discs soaked with 100 ?g of each compound.
Application: Place the prepared discs on the inoculated agar plates. Standards: Chloramphenicol (10 ?g/disc) and Amphotericin-B (100 units/disc) in DMSO.
Incubation:
Refrigerate plates at 8°C for 30 minutes. Incubate at 37°C for 24 hours.
Measurement: Measure the diameters of inhibition zones, including the disc diameter, using a Vernier caliper.
Procedure for Molecular Docking using Schrödinger Glide:
Protein Preparation:
- Protein: E. coli DNA gyrase B (PDB ID: 5L3J). 2) Added hydrogen’s, assigned bond orders, and created zero-order metal bonds.
- Completed missing side chains and loops using Prime.
- Removed water molecules beyond 5 Å from hetero atoms.
- Created het states using Epik.
- Optimized H-bond network to fix redundant hydrogen’s and identify the most probable positions for thiol and hydroxyl hydrogen atoms. 6) Conducted restrained minimization with the OPLS3e force field until RMSD of non-hydrogen atoms converged at 0.30 Å.
Ligands Preparation:
-
- Synthesized compounds (4a-4g) drawn using Chem Bio Draw Ultra.
- Prepared ligands using Schrödinger's Lig Prep for geometry optimization and generating low energy 3D structures with optimal chirality.
Grid Generation:
- Identified the centroid active site based on the co-crystallized ligands using the grid generation tool.
Docking:
- Performed molecular docking using the Glide standard precision (SP) approach.20-22
RESULTS AND DISCUSSION:
Characterization of synthesized compounds:
Compound 4a: IR Spectra:
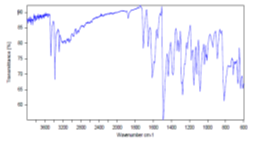
Value: 3372.05 (NH) cm-1, 2859.71 (-CH3), 1711.19 (C=O) cm-1, 1552.12 (C=C) cm-1, 1082.43 (C-O)cm-1
Compound 4b: IR Spectra:

Value: 3371.68 (NH) cm-1, 1737.42 (C=O) cm-1, 1619.85 (C=N) cm-1, 1494.84 (C=C) cm-1, 1202.34 (C-O) cm-1, 707.90 (C-S) cm-1.
compound 4c: IR Spectra:
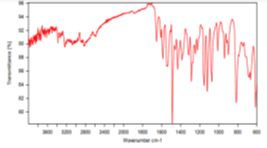
Value: 3378.60 (NH) cm-1, 1735.42 (C=O) cm-1, 1619.85 (C=N) cm-1, 1494.84 (C=C) cm-1, 1202.34 (C-O) cm-1.
compound 4d: IR Spectra:
Value: 3371.68 (NH) cm-1, 1737.42 (C=O) cm-1, 1650.85 (C=N) cm-1, 1490.84 (C=C) cm-1, 1202.34 (C-O) cm-1, 707.90 (C-S) cm-1
compound 4e: IR Spectra:
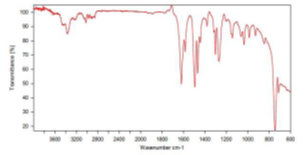
Value: 3380.78 (NH) cm-1, 1711.19 (C=O) cm-1, 1613.50 (C=N) cm-1, 1491.54 (C=C) cm-1, 1222.68 (C-O) cm-1, 708.90 (C-S) cm-1
compound 4f: IR Spectra:

Value: 3371.68 (NH) cm-1, 1737.42 (C=O) cm-1, 1650.85 (C=N) cm-1, 1490.84 (C=C) cm-1, 1202.34 (C-O) cm-1, 707.90 (C-S) cm-1.
compound 4g:IR Spectra:

Value: 3284.15 (NH) cm-1, 1656.55 (C=O) cm-1, 1552.12 (C=N) cm-1, 1434.00 (C=C) cm-1, 1179.60 (C-O) cm-1, 702.90 (C-S) cm-1.
Microbial infections are an escalating threat due to rising antimicrobial resistance. Benzoxazine derivatives have emerged as promising antimicrobial agents due to their diverse biological activities. These compounds, mainly present as aglycones in plants, serve as natural defense chemicals and exhibit anti-inflammatory, antibacterial, and antifungal properties. In this study, 2H-benzo[b][1,4]oxazin-3(4H)-one derivatives were synthesized and evaluated for their antimicrobial efficacy. The synthesis involved reacting 2-aminophenol with chloroacetic acid to form 2H-benzo[b][1,4]oxazin-3(4H)-one, followed by sulfonation with chlorosulfonic acid and subsequent reaction with aryl amines, yielding potent antimicrobial derivatives 4(a-g) shown in fig. no. 3.
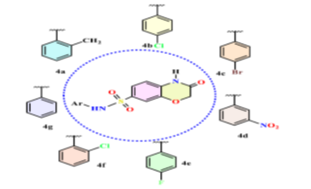
Figure 3: Chemical structure of synthesized compounds.
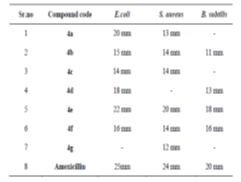
The synthesized compounds were assessed for antimicrobial activity through their zones of inhibition against E. coli, S. aureus, and B. subtilis. Compound 4e emerged as the most potent, showing significant inhibition across all strains: 22 mm for E. coli, 20 mm for S. aureus, and 18 mm for B. subtilis. Compound 4a also showed notable effectiveness against E. coli (20 mm). Compounds 4b, 4c, and 4f displayed moderate activity, while 4g was largely ineffective. These findings highlight compound 4e's potential as a broad-spectrum antimicrobial agent, warranting further in vivo and toxicity studies for clinical application shown in fig no.4 and table no.2.
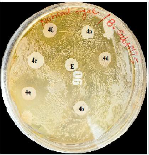
Antimicrobial Activity of compound 4a-4f against B.subtilis.
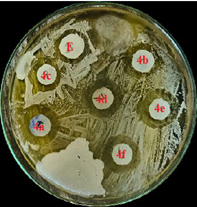
Antimicrobial Activity of compound 4a-4f against E.coli.
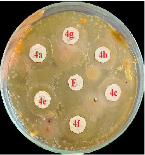
Antimicrobial Activity of compound 4a-4f against S.aureas.
Figure 4: Antimicrobial Activity of compound 4a-4f against various micro organisms.
Table 2: Antibacterial activity of selected synthesized Derivatives:
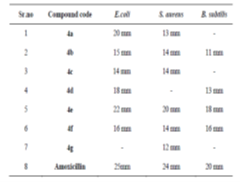
Diameter in mm calculated by Vernier Caliper- means no. zone of inhibition.
Molecular Docking:
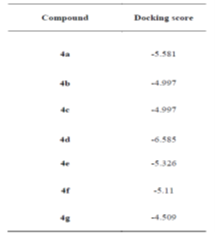
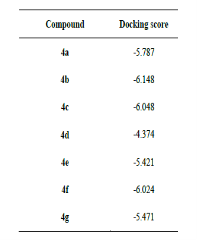
Molecular docking involves in-silico interaction between a macromolecule (protein receptor) and a micro molecule (ligands), often to inhibit the protein's function. DNA gyrase, a type II topoisomerase essential for E. coli, is a primary antibacterial target as its inhibition disrupts bacterial DNA processes. Docking studies for synthesized compounds against E. coli DNA gyrase showed varying binding affinities. Compound 4d had the strongest interaction with a docking score of -6.585. Compounds 4a, 4e, and 4f also demonstrated favorable binding, while compounds 4b, 4c, and 4g showed weaker affinities with higher docking scores.
Table 3: Docking score (in Kcal/mol) of the synthesized compound in active site of E. coli DNA gyrase B (PDB ID: 5L3J)
Using Maestro's ligands-interaction tool, 2D and 3D representations of ligands-protein interactions in fig. 5 and table no. 3 were generated to understand the binding mode of synthesized compounds with E. coli DNA gyrase B. Analysis revealed critical binding residues Met95, Glu50, and Ile59, with compounds 4d, 4a, and 4e forming hydrogen bonds with Asp73 at 2.17Å. These interactions highlight key molecular interactions and inform structure-based drug design. Understanding these interactions enables researchers to optimize compounds for enhanced binding affinity, aiding the development of potent, selective DNA gyrase B inhibitors for therapeutic use.
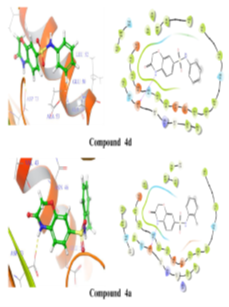
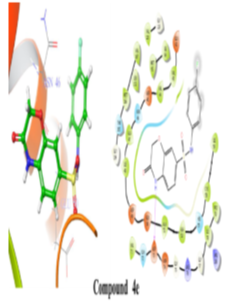
Figure 5: 2D and 3D representation of Binding interaction of active compound 4a and compound 4d in the cavity of E. coli DNA gyrase B (In 2d interaction diagram Magenta arrow indicates hydrogen bonding and green line indicates hydrophobic interaction)
Table 4:Docking score (in Kcal/mol) of the synthesized compound in active site of B.subtilis DNA gyrase B (PDB ID: 6YJ7
The docking results for the synthesized compounds against S. aureus DNA gyrase are summarized in Table 4, with more negative scores indicating stronger interactions. Compound 4a had the strongest interaction with a docking score of -6.038. Compounds 4b, 4c, and 4g also showed favorable binding with low scores. Conversely, compounds 4f, 4d, and 4e exhibited higher docking scores, indicating weaker binding affinities.
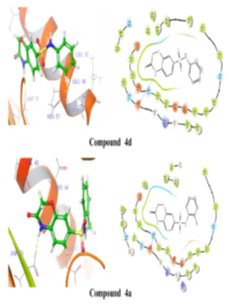
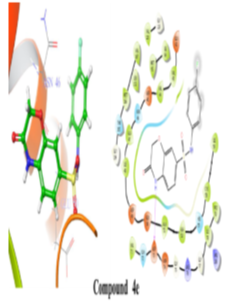
Figure 6: 2D and 3D representation of Binding interaction of active compound 4e in the cavity of E. coli DNA gyrase B (In 2d interaction diagram Magenta arrow indicates hydrogen bonding and green line indicates hydrophobic interaction).
Table 5: Docking score (in Kcal/mol) of the synthesized compound in active site of S.aureas. DNA gyrase B (PDB ID: 3TTZ)
2D and 3D representations of the binding interactions of active compounds 4a and 4g in the cavity of S. aureus DNA gyrase B reveal critical interactions. In the 2D interaction diagrams, magenta arrows indicate hydrogen bonding, while green lines denote hydrophobic interactions. These visualizations help elucidate the binding modes of 4a and 4g within the enzyme's active site, highlighting key interactions that contribute to their binding affinity.
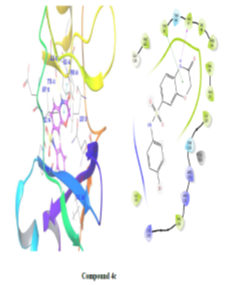
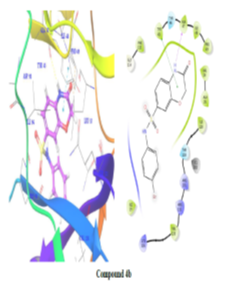
Figure 7: 2D and 3D representation of Binding interaction of active compound 4a and 4g in the cavity of S.aureas. DNA gyrase B (In 2d interaction diagram Magenta arrow indicates hydrogen bonding and green line indicates hydrophobic interaction).
CONCLUTION
In conclusion, the rising prevalence of antimicrobial resistance is a critical global health issue, demanding urgent advancements in therapeutic strategies. With 13.7 million infection-related deaths in 2019, the need for novel antimicrobial agents is evident. Benzoxazine derivatives have emerged as promising candidates due to their diverse biological activities and relative chemical simplicity, offering a versatile approach to combating microbial infections. Our study highlights the potential of these derivatives in addressing infections caused by pathogens such as Staphylococcus aureus, Escherichia coli, Streptococcus pneumonia, Klebsiella pneumonia, and Pseudomonas aeruginosa. Among the synthesized 2H-benzo[b][1,4]oxazin-3(4H)-one derivatives, compound 4e showed the highest antimicrobial potency across all tested strains. Docking studies revealed critical interactions of compounds 4d, 4a, and 4e with the GyrB active site, identifying key amino acid residues essential for binding efficacy. These insights into the molecular mechanisms underlying their antimicrobial activity pave the way for future structure-based drug design. Continued research and development of Benzoxazine derivatives are crucial, offering potential breakthroughs in the fight against microbial infections and antimicrobial resistance. Further studies, including in vivo testing and clinical trials, are essential to validate the efficacy, safety, and therapeutic potential of these compounds. By leveraging our detailed understanding of ligand-protein interactions, researchers can refine Benzoxazine derivatives to develop more potent and selective inhibitors, ultimately improving global health outcomes.
CONFLICT OF INTEREST:
Authors don’t have any conflict of interest
ACKNOWLEDGEMENT:
I would like to express my heartfelt gratitude to ARACOP, including all teaching and non-teaching staff, and Principal Prof. Dr. R.D. Wagh, for their unwavering support throughout this research. I extend special thanks to my research guide, Prof. Dr. R.D. Wagh, for his invaluable guidance and encouragement. Their collective assistance has been instrumental in making this work possible.
REFERENCES
- Bharathkumar H, Sundaram MS, Jagadish S, Paricharak S, Hemshekhar M, Mason D, Kemparaju K, Girish KS, Basappa, Bender A, Rangappa KS. Novel benzoxazine-based aglycones block glucose uptake in vivo by inhibiting glycosidases. PloS one. 2014 Jul 21;9(7):e102759.
- Klun JA, Brindley TA. Role of 6-methoxybenzoxazolinone in inbred resistance of host plant (maize) to first-brood larvae of European corn borer. Journal of economic entomology. 1966 Jun 1;59(3):711-8.
- Argandoña VH, Corcuera LJ, Niemeyer HM, Campbell BC. Toxicity and feeding deterrency of hydroxamic acids from Gramineae in synthetic diets against the greenbug, Schizaphis graminum. Entomologia experimentalis et applicata. 1983 Sep;34(2):134-8.
- Corcuera LJ, Queirolo CB, Argandona VH. Effects of 2-bd-glucosyl-4-hydroxy-7-methoxy-1, 4-benzoxazin-3-one on cereal aphids on artificial diets. Experientia. 1985;41:514-6.
- Toldiné Tóth É. Relationship between DIMBOA content and Helminthosporium turcicum resistance in maize. 1984, 33, 213.
- Coture, R. M.; Roudey, Couture RM, Routley DG, Dunn GM. Role of cyclic hydroxamic acids in monogenic resistance of maize to Helminthosporium turcicum. Physiological Plant Pathology. 1971 Oct 1;1(4):515-21.
- Combs DW, Rampulla MS, Bell SC, Klaubert DH, Tobia AJ, Falotico R, Haertlein B, Lakas-Weiss C, Moore JB. 6-Benzoxazinylpyridazin-3-ones: potent, long-acting positive inotrope and peripheral vasodilator agents. Journal of medicinal chemistry. 1990 Jan;33(1):380-6.
- Li AR, Zhang J, Greenberg J, Lee T, Liu J. Discovery of non-glucoside SGLT2 inhibitors. Bioorganic & medicinal chemistry letters. 2011 Apr 15;21(8):2472-5.
- Iijima, T.; Yamamoto, Y.; Akatsuka, H.; Kawaguchi, T. Worldwide Patent No .200789034. Chem.Abstr. 2007, 147, 257784.
- Wahindulla, S.; Bhattacharjee, J. J. J. Indian. Inst. Sci. 2001, 4, 485.
- Katsura Y, NISHINO S, TAKASUGI H. Studies on antiulcer drugs. I. Synthesis and antiuler activities of imidazo [1, 2-a] pyridinyl-2-oxobenzoxazolidines-3-oxo-2H-1, 4-benzoxazines and related compounds. Chemical and pharmaceutical bulletin. 1991 Nov 25;39(11):2937-43.
- Katritzky AR, Pacureanu LM, Slavov S, Dobchev DA, Karelson M. QSAR study of antiplatelet agents. Bioorganic & medicinal chemistry. 2006 Nov 15;14(22):7490-500.
- KAJINO M, SHIBOUTA Y, NISHIKAWA K, MEGURO K. Synthesis and biological activities of new 2-substituted 1, 4-benzoxazine derivatives. Chemical and pharmaceutical bulletin. 1991 Nov 25;39(11):2896-905.
- Fringuelli R, Pietrella D, Schiaffella F, Guarraci A, Perito S, Bistoni F, Vecchiarelli A. Anti-Candida albicans properties of novel benzoxazine analogues. Bioorganic & medicinal chemistry. 2002 Jun 1;10(6):1681-6.
- Alper-Hayta S, Ak?-Sener E, Tekiner-Gulbas B, Y?ld?z I, Temiz-Arpac? O, Yalc?n I, Altanlar NU. Synthesis, antimicrobial activity and QSARs of new benzoxazine-3-ones. European journal of medicinal chemistry. 2006 Dec 1;41(12):1398-404.
- Deswal S, Roy N. Quantitative structure activity relationship of benzoxazinone derivatives as neuropeptide receptor antagonists. European journal of medicinal chemistry. 2006 Apr 1;41(4):552-7.
- Zhou D, Harrison BL, Shah U, Andree TH, Hornby GA, Scerni R, Schechter LE, Smith DL, Sullivan KM, Mewshaw RE. Studies toward the discovery of the next generation of antidepressants. Part 5: 3, 4-Dihydro-2H-benzo [1, 4] oxazine derivatives with dual 5-HT1A receptor and serotonin transporter affinity. Bioorganic & medicinal chemistry letters. 2006 Mar 1;16(5):1338-41.
- Ohno M, Tanaka Y, Miyamoto M, Takeda T, Hoshi K, Yamada N, Ohtake A. Development of 3, 4-dihydro-2H-benzo [1, 4] oxazine derivatives as dual thromboxane A2 receptor antagonists and prostacyclin receptor agonists. Bioorganic & medicinal chemistry. 2006 Mar 15;14(6):2005-21.
- S. Anderluh P, Anderluh M, Ilas J, Mravljak J, Sollner Dolenc M, Stegnar M, Kikelj D. Toward a novel class of antithrombotic compounds with dual function. Discovery of 1, 4-benzoxazin-3 (4 H)-one derivatives possessing thrombin inhibitory and fibrinogen receptor antagonistic activities. Journal of medicinal chemistry. 2005 May 5;48(9):3110-3.
- Ilas, J.; Tomasic, T.; Kikelj, D. Novel potent and selective thrombin inhibitors based on a central 1, 4-benzoxazin-3 (4 H)-one scaffold. Journal of medicinal chemistry. 2008 May 8;51(9):2863-7.
- Hasui T, Matsunaga N, Ora T, Ohyabu N, Nishigaki N, Imura Y, Igata Y, Matsui H, Motoyaji T, Tanaka T, Habuka N. Identification of benzoxazin-3-one derivatives as novel, potent, and selective nonsteroidal mineralocorticoid receptor antagonists. Journal of medicinal chemistry. 2011 Dec 22;54(24):8616-31.
- Hasui T, Ohra T, Ohyabu N, Asano K, Matsui H, Mizukami A, Habuka N, Sogabe S, Endo S, Siedem CS, Tang TP. Design, synthesis, and structure–activity relationships of dihydrofuran-2-one and dihydropyrrol-2-one derivatives as novel benzoxazin-3-one-based mineralocorticoid receptor antagonists. Bioorganic & medicinal chemistry. 2013 Oct 1;21(19):5983-94.


 Milind Shinde*
Milind Shinde*
 Rajendra D. Wagh
Rajendra D. Wagh





















 10.5281/zenodo.13293096
10.5281/zenodo.13293096